bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6736_orf1
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
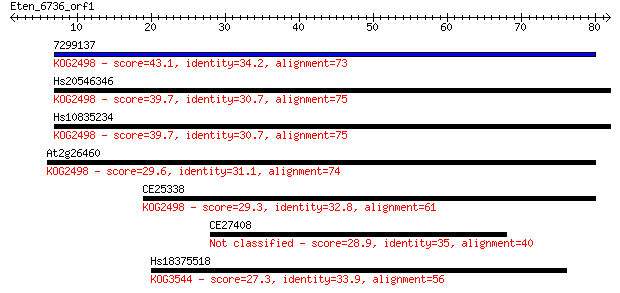
7299137 43.1 1e-04
Hs20546346 39.7 0.001
Hs10835234 39.7 0.001
At2g26460 29.6 1.3
CE25338 29.3 1.9
CE27408 28.9 2.4
Hs18375518 27.3 6.1
> 7299137
Length=557
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 44/75 (58%), Gaps = 4/75 (5%)
Query 7 GAPRGPTRRGPQQFASKAAYEEYMNSIELVPKAAYSYGRRIAGDINSSKNK--KKQKLNP 64
G +GP G F ++ Y +YM++ E +PKAA+ YG ++ + KNK K +K
Sbjct 463 GNKKGPI--GRWDFDTQEEYSDYMSTKEALPKAAFQYGVKMQDGRKTRKNKTEKNEKAEL 520
Query 65 EQEWRAIEKLIAEKK 79
++EW+ I+ +I ++K
Sbjct 521 DREWQKIQTIIQKRK 535
> Hs20546346
Length=557
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 44/76 (57%), Gaps = 3/76 (3%)
Query 7 GAPRGPTRRGPQQFASKAAYEEYMNSIELVPKAAYSYGRRIAGDINSSKNKK-KQKLNPE 65
G +GP G F ++ Y EYMN+ E +PKAA+ YG +++ + + K+ K +
Sbjct 472 GNKKGPL--GRWDFDTQEEYSEYMNNKEALPKAAFQYGIKMSEGRKTRRFKETNDKAELD 529
Query 66 QEWRAIEKLIAEKKQL 81
++W+ I +I ++K++
Sbjct 530 RQWKKISAIIEKRKKM 545
> Hs10835234
Length=557
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 44/76 (57%), Gaps = 3/76 (3%)
Query 7 GAPRGPTRRGPQQFASKAAYEEYMNSIELVPKAAYSYGRRIAGDINSSKNKK-KQKLNPE 65
G +GP G F ++ Y EYMN+ E +PKAA+ YG +++ + + K+ K +
Sbjct 472 GNKKGPL--GRWDFDTQEEYSEYMNNKEALPKAAFQYGIKMSEGRKTRRFKETNDKAELD 529
Query 66 QEWRAIEKLIAEKKQL 81
++W+ I +I ++K++
Sbjct 530 RQWKKISAIIEKRKKM 545
> At2g26460
Length=585
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 6/76 (7%)
Query 6 GGAPRGPTRRGPQQFASKAAYEEYMNSIELVPKAAYSYGRRIAGDINSSKN--KKKQKLN 63
GG +G R F ++ +E+Y E +PKAA+ +G ++ + K + QKLN
Sbjct 490 GGKAKGGLHRW--DFETEEEWEKYNEQKEAMPKAAFQFGVKMQDGRKTRKQNRDRDQKLN 547
Query 64 PEQEWRAIEKLIAEKK 79
E I K++ KK
Sbjct 548 --NELHQINKILTRKK 561
> CE25338
Length=547
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 19 QFASKAAYEEYMNSIELVPKAAYSYGRRIAGDINSSKNKKKQKLNP----EQEWRAIEKL 74
F ++ Y YM E +PKAAY YG + KNKK+ ++ ++E I K+
Sbjct 468 DFDTEEEYASYMEGREALPKAAYQYG--VKNGEGGRKNKKQSAVSDAKRLDRELNEINKI 525
Query 75 IAEKK 79
+ ++K
Sbjct 526 MDKRK 530
> CE27408
Length=433
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 28 EYMNSIELVPKAAYSYGRRIAGDINSSKNKKKQKLNPEQE 67
E IEL+ K A Y +RI DI + + + K++++ QE
Sbjct 251 ESTEKIELIQKEAEDYRKRIEEDIRNRQQRHKKEVDAHQE 290
> Hs18375518
Length=1806
Score = 27.3 bits (59), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 26/62 (41%), Gaps = 6/62 (9%)
Query 20 FASKAAYEEYMNS------IELVPKAAYSYGRRIAGDINSSKNKKKQKLNPEQEWRAIEK 73
F ++ EE I PKAAY Y + D +SS K Q P+ + A E
Sbjct 206 FGTRILDEEVFEGDIQQFLITGDPKAAYDYCEHYSPDCDSSAPKAAQAQEPQIDEYAPED 265
Query 74 LI 75
+I
Sbjct 266 II 267
Lambda K H
0.309 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1162440328
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40