bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6656_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
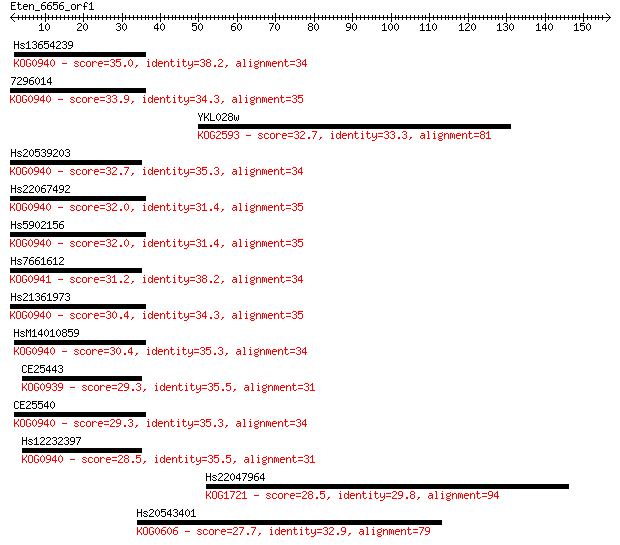
Hs13654239 35.0 0.064
7296014 33.9 0.13
YKL028w 32.7 0.29
Hs20539203 32.7 0.33
Hs22067492 32.0 0.50
Hs5902156 32.0 0.52
Hs7661612 31.2 0.85
Hs21361973 30.4 1.6
HsM14010859 30.4 1.7
CE25443 29.3 3.0
CE25540 29.3 3.3
Hs12232397 28.5 5.5
Hs22047964 28.5 6.2
Hs20543401 27.7 9.1
> Hs13654239
Length=922
Score = 35.0 bits (79), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 2 RPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R V+FRGE D+GG +E+ +++EV+ P+
Sbjct 594 RRLYVIFRGEEGLDYGGLAREWFFLLSHEVLNPM 627
> 7296014
Length=949
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R ++FRGE D+GG +E+ +++EV+ P+
Sbjct 620 RRRLYIIFRGEEGLDYGGVSREWFFLLSHEVLNPM 654
> YKL028w
Length=482
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 16/90 (17%)
Query 50 LPSFNSTHAI---GPHQDSVVLRPDPSAPLPDPLVSASDLLPPALPNTSGGNDGSPSYAE 106
+P N +HA P + S + RP SAPLP +L+ AL N S G+ S A
Sbjct 207 IPPQNQSHAAYTYNPKKGSTMFRPGDSAPLP-------NLMGTALGNDSSRRAGANSQAT 259
Query 107 V------SPDTVTQEPVQSVGSQPDRSSHA 130
+ + D V Q +Q ++ R +A
Sbjct 260 LHINITTASDEVAQRELQERQAEEKRKQNA 289
> Hs20539203
Length=757
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGP 34
++ +V FRGE D+GG +E+L + +E++ P
Sbjct 425 KKRLMVKFRGEEGLDYGGVAREWLYLLCHEMLNP 458
> Hs22067492
Length=870
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R ++ RGE D+GG +E+ +++EV+ P+
Sbjct 541 RRRLYIIMRGEEGLDYGGIAREWFFLLSHEVLNPM 575
> Hs5902156
Length=870
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R ++ RGE D+GG +E+ +++EV+ P+
Sbjct 541 RRRLYIIMRGEEGLDYGGIAREWFFLLSHEVLNPM 575
> Hs7661612
Length=110
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGP 34
++P V+F GE A D GG +EF + E++ P
Sbjct 52 KKPLKVIFVGEDAVDAGGVRKEFFLLIMRELLDP 85
> Hs21361973
Length=903
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 1 ERPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R V+F GE D+GG +E+ +++EV+ P+
Sbjct 574 RRRLWVIFPGEEGLDYGGVAREWFFLLSHEVLNPM 608
> HsM14010859
Length=862
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 2 RPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R V+F GE D+GG +E+ +++EV+ P+
Sbjct 534 RRLWVIFPGEEGLDYGGVAREWFFLLSHEVLNPM 567
> CE25443
Length=875
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 4 FLVVFRGEGATDFGGPFQEFLSGVANEVMGP 34
F ++F+GE D GG +E+ S + E+ P
Sbjct 547 FYIIFQGEEGQDAGGLLREWFSVITREIFNP 577
> CE25540
Length=817
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 2 RPFLVVFRGEGATDFGGPFQEFLSGVANEVMGPL 35
R + FRGE D+GG +E+ +++EV+ P+
Sbjct 466 RRLYIQFRGEEGLDYGGVAREWFFLLSHEVLNPM 499
> Hs12232397
Length=748
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 4 FLVVFRGEGATDFGGPFQEFLSGVANEVMGP 34
++ FRGE D+GG +E+L +++E++ P
Sbjct 422 LMIKFRGEEGLDYGGVAREWLYLLSHEMLNP 452
> Hs22047964
Length=1102
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 38/97 (39%), Gaps = 18/97 (18%)
Query 52 SFNSTHAIGPHQDSVVLR---PDPSAPLPDPLVSASDLLPPALPNTSGGNDGSPSYAEVS 108
S + +G Q+ VL PDPS L ASDLLPP +GG
Sbjct 635 SHGAQSPLGEGQNMAVLSAGDPDPSRCLRSNPAEASDLLPP----VAGGG---------- 680
Query 109 PDTVTQEPVQSVGSQPDRSSHAEFGEGPQSIRSNLSN 145
DT+T +P + RS F + S++ N
Sbjct 681 -DTITHQPDSCKAAPEHRSGITAFMKVLNSLQKKQMN 716
> Hs20543401
Length=2092
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 38/85 (44%), Gaps = 6/85 (7%)
Query 34 PLAESPDE--LLPTFLACLPSFNSTHAI----GPHQDSVVLRPDPSAPLPDPLVSASDLL 87
P+ +SP E L ++CL T I GP L SA P P +
Sbjct 1249 PVRKSPSEYKLEGRSVSCLKPIEGTLDIALLSGPQASKTELPSPESAQSPSPSGDVRASV 1308
Query 88 PPALPNTSGGNDGSPSYAEVSPDTV 112
PP LP++SG + + S E+SP ++
Sbjct 1309 PPVLPSSSGKKNDTTSARELSPSSL 1333
Lambda K H
0.311 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40