bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6652_orf2
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
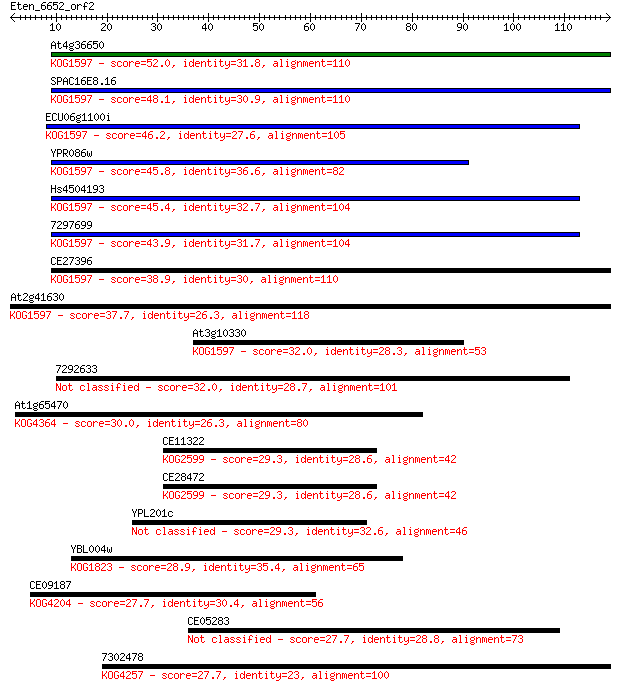
At4g36650 52.0 2e-07
SPAC16E8.16 48.1 4e-06
ECU06g1100i 46.2 2e-05
YPR086w 45.8 2e-05
Hs4504193 45.4 2e-05
7297699 43.9 7e-05
CE27396 38.9 0.003
At2g41630 37.7 0.006
At3g10330 32.0 0.25
7292633 32.0 0.29
At1g65470 30.0 1.1
CE11322 29.3 1.7
CE28472 29.3 1.8
YPL201c 29.3 2.1
YBL004w 28.9 2.7
CE09187 27.7 5.1
CE05283 27.7 5.6
7302478 27.7 5.6
> At4g36650
Length=503
Score = 52.0 bits (123), Expect = 2e-07, Method: Composition-based stats.
Identities = 35/111 (31%), Positives = 58/111 (52%), Gaps = 11/111 (9%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
V++KE+GK I + + L IN++ + H+ PR+C+ LQL+ ++A H+ +
Sbjct 202 VQQKEIGKYIKILGEALQLSQPINSNSISVHM-PRFCTLLQLNKSAQELATHIGEVVINK 260
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEHT 118
C+ R P S++AAAI+L QL + AE+ +TG E T
Sbjct 261 CFCTRRNPISISAAAIYLACQLEDKRKTQ---------AEICKITGLTEVT 302
> SPAC16E8.16
Length=340
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 61/125 (48%), Gaps = 24/125 (19%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNS--------------ESASHLLPRYCSKLQLSVHV 54
V +KE+G+ ++++L G ++NS SA L+ R+C++L L + V
Sbjct 190 VPKKEIGRVYKTLQRMLTEGGALHNSVDALKGHEYIQSSSTSAEDLMVRFCNRLMLPMSV 249
Query 55 SDVAEHVAKRAA-QVFVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTG 113
A +A+RA Q + P S+AA+ I+++ L+ + PK E++ VTG
Sbjct 250 QSAAAELARRAGLQGTLAGRSPISIAASGIYMISALMGYPKT-------PK--EISEVTG 300
Query 114 AGEHT 118
+ T
Sbjct 301 VSDST 305
> ECU06g1100i
Length=312
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 54/106 (50%), Gaps = 7/106 (6%)
Query 8 AVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQ 67
AV+++E+G+ I L + S +++ R+CS L L++ + +A +AK A +
Sbjct 181 AVQKREIGRCFKLISPHLERMATM----STENIIARFCSDLNLNIKIQKIATEIAKAAHE 236
Query 68 VFVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ + + P+S+AAA I++V L Q+ + V V VT
Sbjct 237 LGCLAGKSPDSIAAAVIYMVTNLFPEEKKIQKDIQF--VTNVTEVT 280
> YPR086w
Length=345
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 49/94 (52%), Gaps = 14/94 (14%)
Query 9 VKEKELGKQINKIKKLLPTRG---------GINNSESASHL--LPRYCSKLQLSVHVSDV 57
VK KE GK +N +K +L RG +N A +L +PR+CS L L + V+
Sbjct 198 VKTKEFGKTLNIMKNIL--RGKSEDGFLKIDTDNMSGAQNLTYIPRFCSHLGLPMQVTTS 255
Query 58 AEHVAKRAAQVF-VCSHRPNSVAAAAIWLVVQLL 90
AE+ AK+ ++ + P ++A +I+L + L
Sbjct 256 AEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF 289
> Hs4504193
Length=316
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 55/105 (52%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L V A H+A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETSVDL---ITTGDFMSRFCSNLCLPKQVQMAATHIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKRTQKEIGDIAGVADVT 284
> 7297699
Length=315
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ K L T + + + + R+C+ L L V A H+AK+A ++
Sbjct 185 ISKKEIGRCFKLTLKALETSVDL---ITTADFMCRFCANLDLPNMVQRAATHIAKKAVEM 241
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S + Q + +A VA VT
Sbjct 242 DIVPGRSPISVAAAAIYMASQ---ASEHKRSQKEIGDIAGVADVT 283
> CE27396
Length=306
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 52/113 (46%), Gaps = 17/113 (15%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSE--SASHLLPRYCSKLQLSVHVSDVAEHVAKRAA 66
V +KE+G+ I + L T N E +++ + R+C L L + A +AK A
Sbjct 176 VSKKEIGRCFKIIVRSLET-----NLEQITSADFMSRFCGNLSLPNSIQAAATRIAKCAV 230
Query 67 QV-FVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEHT 118
+ V P S+AAAAI++ Q + S++ E+ V GA E T
Sbjct 231 DMDLVAGRTPISIAAAAIYMASQASAEKRSAK---------EIGDVAGAAEIT 274
> At2g41630
Length=312
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 55/121 (45%), Gaps = 13/121 (10%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGIN---NSESASHLLPRYCSKLQLSVHVSDV 57
E+ V +KE+G+ + I K L G + + A + R+CS L +S H
Sbjct 170 EICVIANGATKKEIGRAKDYIVKTLGLEPGQSVDLGTIHAGDFMRRFCSNLAMSNHAVKA 229
Query 58 AEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEH 117
A+ +++ + F P S+AA I+++ QL S ++ L +++ TG E
Sbjct 230 AQEAVQKSEE-FDIRRSPISIAAVVIYIITQL----SDDKKTL-----KDISHATGVAEG 279
Query 118 T 118
T
Sbjct 280 T 280
> At3g10330
Length=312
Score = 32.0 bits (71), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 37 ASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQL 89
A + R+CS L ++ A+ +++ + F P S+AAA I+++ QL
Sbjct 209 AGDFMRRFCSNLGMTNQTVKAAQESVQKSEE-FDIRRSPISIAAAVIYIITQL 260
> 7292633
Length=943
Score = 32.0 bits (71), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 49/108 (45%), Gaps = 27/108 (25%)
Query 10 KEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVF 69
+E++ K++ ++++LPT GGI+++ S P+Y + H D R +
Sbjct 729 QEQQRDKELTTLREILPTGGGIDDANS-----PKYFPPETTTFHEED------GRVVKKI 777
Query 70 VCSHRPNSVAAAAIWLVV----QLLSSSSSSQQ---QLALPKVAEVAS 110
C W VV +LS + Q+ +LAL K+A VAS
Sbjct 778 TCYE---------TWHVVSTAKDMLSKMARQQRTCLELALVKLANVAS 816
> At1g65470
Length=815
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 41/83 (49%), Gaps = 5/83 (6%)
Query 2 LVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESA-SHLLPRYCSKLQLSVHVSDVAEH 60
+V D + E E G Q++++ + P+ N + S P +C+ LQ H+ ++ +H
Sbjct 563 FMVPDGYLSEDE-GVQVDRMD-IDPSEQDANTTSSKQDQESPEFCALLQQQKHLQNLTDH 620
Query 61 VAKRAAQVFVC--SHRPNSVAAA 81
K+ + +C +H S+ AA
Sbjct 621 ALKKTQPLIICNLTHEKVSLLAA 643
> CE11322
Length=348
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 31 INNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQ-VFVCS 72
+NN + +H+L YC + ++DV + + K+ FVC
Sbjct 119 LNNINNYTHVLTGYCGNVTFLQKIADVVKDLKKKNGNTTFVCD 161
> CE28472
Length=338
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 31 INNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQ-VFVCS 72
+NN + +H+L YC + ++DV + + K+ FVC
Sbjct 109 LNNINNYTHVLTGYCGNVTFLQKIADVVKDLKKKNGNTTFVCD 151
> YPL201c
Length=461
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 23/51 (45%), Gaps = 5/51 (9%)
Query 25 LPTRGGINNSESA-----SHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFV 70
+PTR I NS S+ S + P + +L VH S +A +F+
Sbjct 341 IPTRDPIENSNSSPPVSDSEVYPIFYKTQELHVHASGTGRQIANNGKYIFI 391
> YBL004w
Length=2493
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query 13 ELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDV-AEHVAKRAAQVFVC 71
E G ++ +K LL R + N S LL RY L H SD +E + K Q+F
Sbjct 1765 EFGTLLSPVKALLMVRINLRNQNKLSELLRRYLLGLN---HNSDSESESILKFCHQLFQE 1821
Query 72 SHRPNS 77
S NS
Sbjct 1822 SEMSNS 1827
> CE09187
Length=1490
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Query 5 YD-RAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
YD +A K K L +N+I+++ R N+++++ HL+ Y + ++ V+DV H
Sbjct 989 YDQKAFKSKPL---VNQIEQICEERRKNNSTDTSPHLILEYTPERKVYRDVNDVTGH 1042
> CE05283
Length=1058
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 36 SASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSS 95
SA+ L C+K Q+ + V DV +HV + + PN + L L+ ++S
Sbjct 424 SANEFLITVCAKNQVPLFV-DVFDHVQQAGDIRNIFGQNPNLKRSIGRQLAQHLIKTASP 482
Query 96 SQQQLALPKVAEV 108
S++ + KVA V
Sbjct 483 SEKAALIAKVASV 495
> 7302478
Length=1200
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 41/100 (41%), Gaps = 9/100 (9%)
Query 19 NKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSV 78
N ++ L+ T GIN +ES + L+ YC L + + H A+ PN
Sbjct 353 NNVEDLVITCNGINTAESIADLIDGYCRLLSKDLEFT--IWHRETNASNEDSAKALPNDA 410
Query 79 AAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEHT 118
+ S+SS + + AE+ + G G+++
Sbjct 411 TLGSN-------KSTSSQGKPMLTDDYAEIGLLEGEGDYS 443
Lambda K H
0.313 0.125 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40