bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6597_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
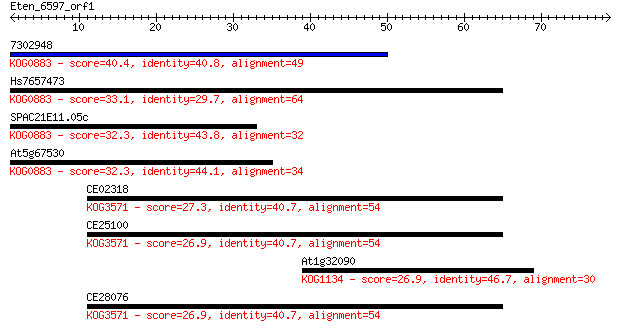
7302948 40.4 8e-04
Hs7657473 33.1 0.12
SPAC21E11.05c 32.3 0.20
At5g67530 32.3 0.23
CE02318 27.3 6.9
CE25100 26.9 8.2
At1g32090 26.9 8.3
CE28076 26.9 9.7
> 7302948
Length=517
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 1 GGLQVLRAWETIPVDSRDRPQRPLFIQKMHIFKNGIEEAKLKLEKEKNE 49
GGL L+ E I VD++DRP + I+ +F N EA +L KE+ E
Sbjct 402 GGLDTLQKMENIEVDNKDRPIEDIIIESSQVFVNPFAEAAEQLAKEREE 450
> Hs7657473
Length=520
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 1/65 (1%)
Query 1 GGLQVLRAWETIPVDSR-DRPQRPLFIQKMHIFKNGIEEAKLKLEKEKNEKLNRQAEEAK 59
GG VL A E + D + DRP+ + I +F + EEA ++ +E+ +L E
Sbjct 402 GGFDVLTAMENVESDPKTDRPKEEIRIDATTVFVDPYEEADAQIAQERKTQLKVAPETKV 461
Query 60 AQARP 64
++P
Sbjct 462 KSSQP 466
> SPAC21E11.05c
Length=471
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 1 GGLQVLRAWETIPVDSRDRPQRPLFIQKMHIF 32
GGL VL A E +P +S D P+ P+ ++ + IF
Sbjct 353 GGLNVLDALEKVPTNSNDHPKLPIKLEDIIIF 384
> At5g67530
Length=595
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 1 GGLQVLRAWETIPVDSRDRPQRPLFIQKMHIFKN 34
GGL L A E +PVD DRP + I + +F N
Sbjct 466 GGLATLAAMENVPVDESDRPLEEIKIIEASVFVN 499
> CE02318
Length=672
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 9/63 (14%)
Query 11 TIPVDS--RDRPQRPLFIQKMH-IFKNGIEEAKLKLEKEKNEKLNRQAEE------AKAQ 61
T+P S D +R L+ Q+ + I + IEEA+ K E E+NEK+ + E ++
Sbjct 365 TLPFTSTASDDEERMLYDQRRNGIPRALIEEAERKRENEQNEKIEQLTEMIDPIIVVRSM 424
Query 62 ARP 64
ARP
Sbjct 425 ARP 427
> CE25100
Length=666
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 9/63 (14%)
Query 11 TIPVDS--RDRPQRPLFIQKMH-IFKNGIEEAKLKLEKEKNEKLNRQAEE------AKAQ 61
T+P S D +R L+ Q+ + I + IEEA+ K E E+NEK+ + E ++
Sbjct 365 TLPFTSTASDDEERMLYDQRRNGIPRALIEEAERKRENEQNEKIEQLTEMIDPIIVVRSM 424
Query 62 ARP 64
ARP
Sbjct 425 ARP 427
> At1g32090
Length=806
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 39 AKLKLEKEKNEKLNRQAEEAKAQARPWFNN 68
AK KLEKE +LN +A+ A A P F++
Sbjct 689 AKDKLEKETEPELNMKADLADAYLHPIFHS 718
> CE28076
Length=625
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 9/63 (14%)
Query 11 TIPVDS--RDRPQRPLFIQKMH-IFKNGIEEAKLKLEKEKNEKLNRQAEE------AKAQ 61
T+P S D +R L+ Q+ + I + IEEA+ K E E+NEK+ + E ++
Sbjct 324 TLPFTSTASDDEERMLYDQRRNGIPRALIEEAERKRENEQNEKIEQLTEMIDPIIVVRSM 383
Query 62 ARP 64
ARP
Sbjct 384 ARP 386
Lambda K H
0.316 0.132 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171925608
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40