bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6593_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
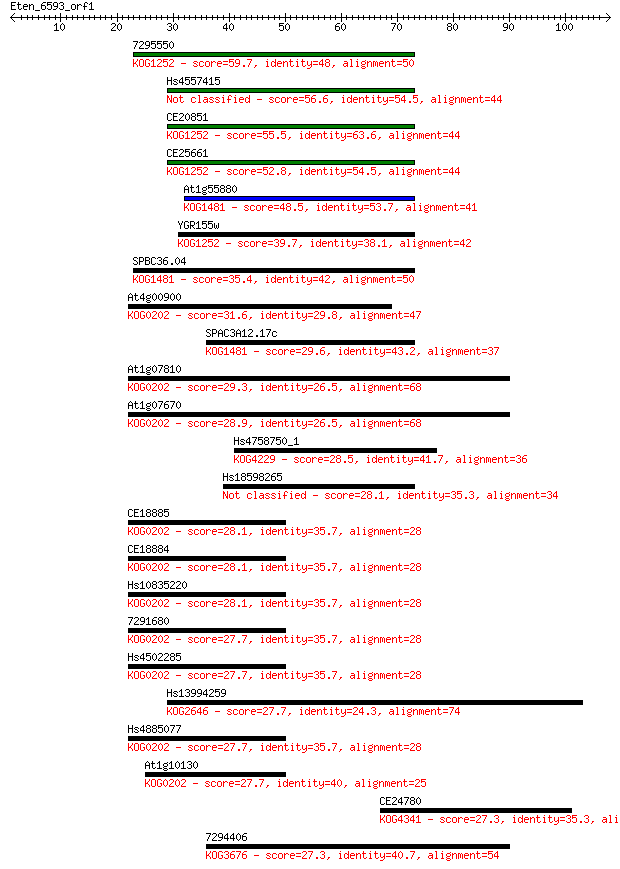
7295550 59.7 1e-09
Hs4557415 56.6 1e-08
CE20851 55.5 2e-08
CE25661 52.8 1e-07
At1g55880 48.5 3e-06
YGR155w 39.7 0.001
SPBC36.04 35.4 0.025
At4g00900 31.6 0.40
SPAC3A12.17c 29.6 1.3
At1g07810 29.3 2.1
At1g07670 28.9 2.1
Hs4758750_1 28.5 3.3
Hs18598265 28.1 3.7
CE18885 28.1 4.3
CE18884 28.1 4.4
Hs10835220 28.1 4.5
7291680 27.7 4.8
Hs4502285 27.7 5.2
Hs13994259 27.7 5.4
Hs4885077 27.7 5.9
At1g10130 27.7 6.0
CE24780 27.3 6.1
7294406 27.3 6.3
> 7295550
Length=522
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 24/50 (48%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 23 SHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
+H + P+IL+ IGCTPLV L+ + + G+ C++ KCEFL+ GGS+KD
Sbjct 40 AHRQQITPNILEVIGCTPLVKLNNIPASDGIECEMYAKCEFLNPGGSVKD 89
> Hs4557415
Length=551
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/44 (54%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 29 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
LP IL IG TP+V ++++G G+ C+LL KCEF +AGGS+KD
Sbjct 77 LPDILKKIGDTPMVRINKIGKKFGLKCELLAKCEFFNAGGSVKD 120
> CE20851
Length=755
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/44 (63%), Positives = 31/44 (70%), Gaps = 0/44 (0%)
Query 29 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
L S+LDAIG TPLV L V A GV C + KCEFL+AGGS KD
Sbjct 337 LDSVLDAIGKTPLVKLQHVPKAHGVRCNVYVKCEFLNAGGSTKD 380
> CE25661
Length=700
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/44 (54%), Positives = 31/44 (70%), Gaps = 0/44 (0%)
Query 29 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
L ++LDAIG TPLV L + A GV C + KCE+++AGGS KD
Sbjct 375 LDTVLDAIGKTPLVKLQHIPKAHGVKCNVYVKCEYMNAGGSTKD 418
> At1g55880
Length=421
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 22/41 (53%), Positives = 32/41 (78%), Gaps = 2/41 (4%)
Query 32 ILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
++DAIG TPL+ ++ + A G C++LGKCEFL+ GGS+KD
Sbjct 44 LVDAIGNTPLIRINSLSEATG--CEILGKCEFLNPGGSVKD 82
> YGR155w
Length=507
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 31 SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
+++D +G TPL+ L ++ A G+ Q+ K E + GGS+KD
Sbjct 13 NVIDLVGNTPLIALKKLPKALGIKPQIYAKLELYNPGGSIKD 54
> SPBC36.04
Length=351
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 31/54 (57%), Gaps = 6/54 (11%)
Query 23 SHLARELPSILD----AIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
S + ++P I+ AIG TPL+ L+ + G C +L K EF + GGS+KD
Sbjct 4 SGIQTKVPGIVSGFIGAIGRTPLIRLNTLSNETG--CNILAKAEFQNPGGSVKD 55
> At4g00900
Length=1054
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 28/47 (59%), Gaps = 4/47 (8%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGG 68
++ + R+LPS+ + +GCT ++C + GT ++ + EF + GG
Sbjct 346 KNAIVRKLPSV-ETLGCTTVICSDKTGT---LTTNQMSATEFFTLGG 388
> SPAC3A12.17c
Length=395
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 36 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
IG T +V + + A G C +L K EFL+ G S KD
Sbjct 50 IGNTKMVRIKSLSQATG--CDILAKAEFLNPGNSPKD 84
> At1g07810
Length=1061
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 8/72 (11%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAG---GSMKDMKRNGC 78
++ L R+LPS+ + +GCT ++C + GT ++ + + ++ G G+++ G
Sbjct 361 KNALVRKLPSV-ETLGCTTVICSDKTGT---LTTNQMAVSKLVAMGSRIGTLRSFNVEGT 416
Query 79 GNDHRS-KLENF 89
D R K+E++
Sbjct 417 SFDPRDGKIEDW 428
> At1g07670
Length=1061
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 8/72 (11%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAG---GSMKDMKRNGC 78
++ L R+LPS+ + +GCT ++C + GT ++ + + ++ G G+++ G
Sbjct 361 KNALVRKLPSV-ETLGCTTVICSDKTGT---LTTNQMAVSKLVAMGSRIGTLRSFNVEGT 416
Query 79 GNDHRS-KLENF 89
D R K+E++
Sbjct 417 SFDPRDGKIEDW 428
> Hs4758750_1
Length=1144
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 41 LVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDMKRN 76
L LS+ G A+GV +LG C L A G+ K N
Sbjct 256 LTALSQKGYASGVERTILGACPVLEAFGNAKTAHNN 291
> Hs18598265
Length=292
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 39 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 72
TP +C S+VG + + C + G + +AGG+ +D
Sbjct 256 TPTICGSQVGKSHHLLCWMPGPPSWGTAGGARED 289
> CE18885
Length=1004
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 331 KNAIVRSLPSV-ETLGCTSVICSDKTGT 357
> CE18884
Length=1059
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 331 KNAIVRSLPSV-ETLGCTSVICSDKTGT 357
> Hs10835220
Length=994
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 329 KNAIVRSLPSV-ETLGCTSVICSDKTGT 355
> 7291680
Length=1020
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 329 KNAIVRSLPSV-ETLGCTSVICSDKTGT 355
> Hs4502285
Length=997
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 329 KNAIVRSLPSV-ETLGCTSVICSDKTGT 355
> Hs13994259
Length=430
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 18/76 (23%), Positives = 33/76 (43%), Gaps = 2/76 (2%)
Query 29 LPSILDAIGCTPLVCLSRVGTAAGVSCQL--LGKCEFLSAGGSMKDMKRNGCGNDHRSKL 86
+ + + A+GC P++C G G C L L L+ + + + G
Sbjct 1 MATAVRAVGCLPVLCSGTAGHLLGRQCSLNTLPAASILAWKSVLGNGHLSSLGTRDTHPY 60
Query 87 ENFSQSLESLKYISSP 102
+ S++L++ ISSP
Sbjct 61 ASLSRALQTQCCISSP 76
> Hs4885077
Length=999
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 22 QSHLARELPSILDAIGCTPLVCLSRVGT 49
++ + R LPS+ + +GCT ++C + GT
Sbjct 329 KNAIVRSLPSV-ETLGCTSVICSDKTGT 355
> At1g10130
Length=992
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 17/25 (68%), Gaps = 1/25 (4%)
Query 25 LARELPSILDAIGCTPLVCLSRVGT 49
+ R LPS+ + +GCT ++C + GT
Sbjct 322 IVRSLPSV-ETLGCTTVICSDKTGT 345
> CE24780
Length=461
Score = 27.3 bits (59), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 67 GGSMKDMKRNGCGNDHRSKLENFSQSLESLKYIS 100
GG +K++ GC N H S L F+ +L+++S
Sbjct 122 GGFLKELSLKGCENVHDSALRTFTSRCPNLEHLS 155
> 7294406
Length=750
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 28/62 (45%), Gaps = 8/62 (12%)
Query 36 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDMKRNGCGN------DH--RSKLE 87
+G T S V AGVS Q L K L GG + DM + C DH ++K+E
Sbjct 1 MGNTESNVTSGVKKQAGVSTQALYKFVNLKGGGLLVDMMKRACQTKQFAEIDHAIKTKVE 60
Query 88 NF 89
F
Sbjct 61 PF 62
Lambda K H
0.318 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40