bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6549_orf2
Length=115
Score E
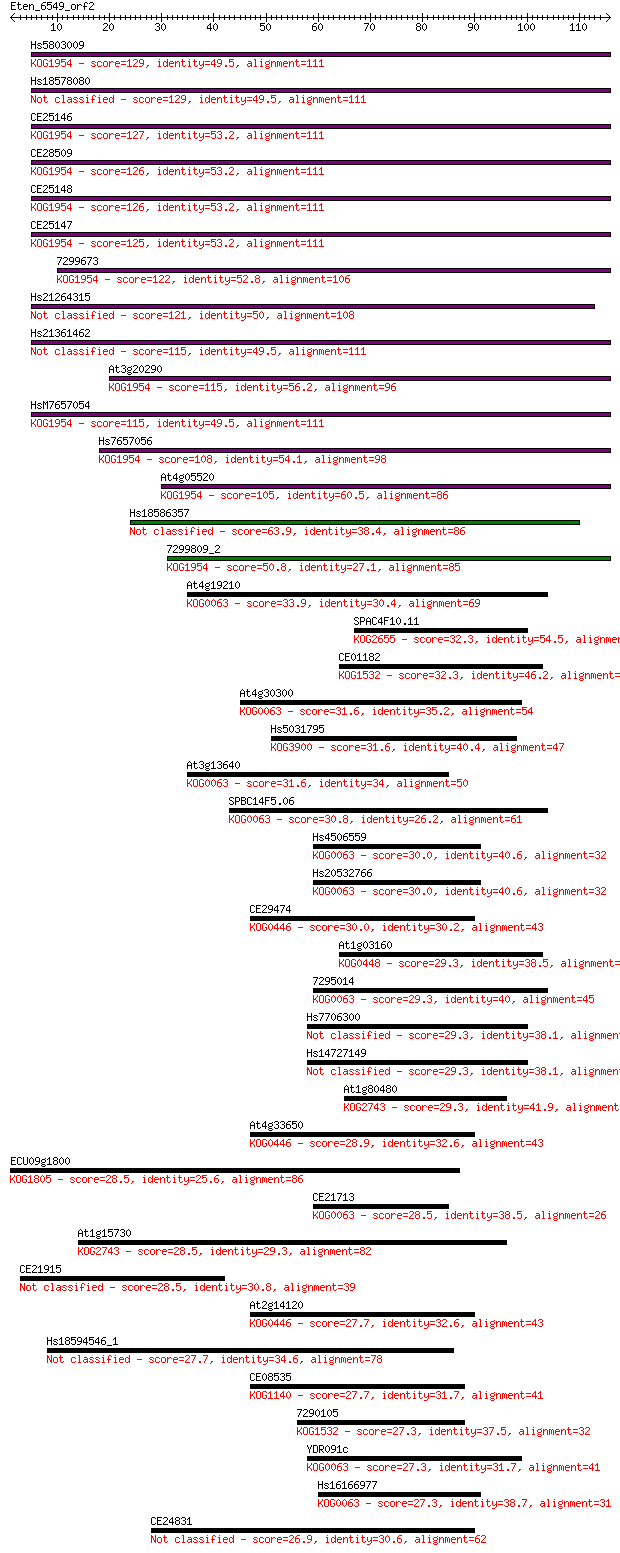
Sequences producing significant alignments: (Bits) Value
Hs5803009 129 2e-30
Hs18578080 129 2e-30
CE25146 127 5e-30
CE28509 126 1e-29
CE25148 126 1e-29
CE25147 125 2e-29
7299673 122 2e-28
Hs21264315 121 2e-28
Hs21361462 115 1e-26
At3g20290 115 2e-26
HsM7657054 115 2e-26
Hs7657056 108 3e-24
At4g05520 105 1e-23
Hs18586357 63.9 8e-11
7299809_2 50.8 7e-07
At4g19210 33.9 0.078
SPAC4F10.11 32.3 0.20
CE01182 32.3 0.23
At4g30300 31.6 0.33
Hs5031795 31.6 0.38
At3g13640 31.6 0.42
SPBC14F5.06 30.8 0.64
Hs4506559 30.0 0.95
Hs20532766 30.0 0.98
CE29474 30.0 1.1
At1g03160 29.3 1.6
7295014 29.3 1.7
Hs7706300 29.3 1.8
Hs14727149 29.3 1.8
At1g80480 29.3 2.0
At4g33650 28.9 2.3
ECU09g1800 28.5 2.8
CE21713 28.5 2.8
At1g15730 28.5 2.8
CE21915 28.5 3.3
At2g14120 27.7 4.8
Hs18594546_1 27.7 5.2
CE08535 27.7 5.6
7290105 27.3 6.6
YDR091c 27.3 7.7
Hs16166977 27.3 7.8
CE24831 26.9 8.8
> Hs5803009
Length=534
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 82/111 (73%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M W+ S+ + ++ ++ V GL+ +Y + LLPLE ++F +F+SP L D DF K
Sbjct 1 MFSWV--SKDARRKKEPELFQTVAEGLRQLYAQKLLPLEEHYRFHEFHSPALEDADFDNK 58
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PMV+++GQYSTGKTTF++HL+E+D+PG+RIGPEPTTD F+A+M G T+ V+
Sbjct 59 PMVLLVGQYSTGKTTFIRHLIEQDFPGMRIGPEPTTDSFIAVMHGPTEGVV 109
> Hs18578080
Length=534
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 82/111 (73%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M W+ S+ + ++ ++ V GL+ +Y + LLPLE ++F +F+SP L D DF K
Sbjct 1 MFSWV--SKDARRKKEPELFQTVAEGLRQLYAQKLLPLEEHYRFHEFHSPALEDADFDNK 58
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PMV+++GQYSTGKTTF++HL+E+D+PG+RIGPEPTTD F+A+M G T+ V+
Sbjct 59 PMVLLVGQYSTGKTTFIRHLIEQDFPGMRIGPEPTTDSFIAVMHGPTEGVV 109
> CE25146
Length=786
Score = 127 bits (319), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 79/111 (71%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL G S +++ V V GL+ +YK+ LLPLE KF F+SP L D DF AK
Sbjct 232 MFSWLGGD--SSKKKNKEVLETVSEGLRKIYKQKLLPLEEFHKFHDFHSPALDDPDFDAK 289
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PM++++GQYSTGKTTF+++LLE D+PG+RIGPEPTTD+F+A+M G + I
Sbjct 290 PMILLVGQYSTGKTTFIRYLLESDFPGIRIGPEPTTDRFIAVMHGDEEGSI 340
> CE28509
Length=571
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 79/111 (71%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL G S +++ V V GL+ +YK+ LLPLE KF F+SP L D DF AK
Sbjct 17 MFSWLGGD--SSKKKNKEVLETVSEGLRKIYKQKLLPLEEFHKFHDFHSPALDDPDFDAK 74
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PM++++GQYSTGKTTF+++LLE D+PG+RIGPEPTTD+F+A+M G + I
Sbjct 75 PMILLVGQYSTGKTTFIRYLLESDFPGIRIGPEPTTDRFIAVMHGDEEGSI 125
> CE25148
Length=589
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 79/111 (71%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL G S +++ V V GL+ +YK+ LLPLE KF F+SP L D DF AK
Sbjct 35 MFSWLGGD--SSKKKNKEVLETVSEGLRKIYKQKLLPLEEFHKFHDFHSPALDDPDFDAK 92
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PM++++GQYSTGKTTF+++LLE D+PG+RIGPEPTTD+F+A+M G + I
Sbjct 93 PMILLVGQYSTGKTTFIRYLLESDFPGIRIGPEPTTDRFIAVMHGDEEGSI 143
> CE25147
Length=835
Score = 125 bits (314), Expect = 2e-29, Method: Composition-based stats.
Identities = 59/111 (53%), Positives = 79/111 (71%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL G S +++ V V GL+ +YK+ LLPLE KF F+SP L D DF AK
Sbjct 281 MFSWLGGD--SSKKKNKEVLETVSEGLRKIYKQKLLPLEEFHKFHDFHSPALDDPDFDAK 338
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PM++++GQYSTGKTTF+++LLE D+PG+RIGPEPTTD+F+A+M G + I
Sbjct 339 PMILLVGQYSTGKTTFIRYLLESDFPGIRIGPEPTTDRFIAVMHGDEEGSI 389
> 7299673
Length=493
Score = 122 bits (305), Expect = 2e-28, Method: Composition-based stats.
Identities = 56/106 (52%), Positives = 78/106 (73%), Gaps = 3/106 (2%)
Query 10 FGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMV 69
F R+ TQ+ V +V L+ +Y+ LLPLE ++F F+SP L D DF AKPM+++
Sbjct 4 FLKREKNTQE---VVENVIGELKKIYRSKLLPLEEHYQFHDFHSPKLEDPDFDAKPMILL 60
Query 70 LGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
+GQYSTGKTTF+++LLERD+PG+RIGPEPTTD+F+A+M + VI
Sbjct 61 VGQYSTGKTTFIRYLLERDFPGIRIGPEPTTDRFIAVMYDDKEGVI 106
> Hs21264315
Length=541
Score = 121 bits (304), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 54/110 (49%), Positives = 81/110 (73%), Gaps = 3/110 (2%)
Query 5 MSRWLFGSRKSGTQQS--ASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFA 62
M W+ G + G +++ A V GL+++Y + +LPLE ++F +F+SP L D DF
Sbjct 1 MFSWM-GRQAGGRERAGGADAVQTVTGGLRSLYLRKVLPLEEAYRFHEFHSPALEDADFE 59
Query 63 AKPMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTD 112
KPM++++GQYSTGKTTF+++LLE+D+PG+RIGPEPTTD F+A+M G T+
Sbjct 60 NKPMILLVGQYSTGKTTFIRYLLEQDFPGMRIGPEPTTDSFIAVMYGETE 109
> Hs21361462
Length=543
Score = 115 bits (289), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 72/111 (64%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL R Q V + L+ +Y+ LLPLE ++F F+SP L D DF K
Sbjct 1 MFSWL--KRGGARGQQPEAIRTVTSALKELYRTKLLPLEEHYRFGAFHSPALEDADFDGK 58
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PMV+V GQYSTGKT+F+Q+LLE++ PG R+GPEPTTD FVA+M G T+ +
Sbjct 59 PMVLVAGQYSTGKTSFIQYLLEQEVPGSRVGPEPTTDCFVAVMHGDTEGTV 109
> At3g20290
Length=485
Score = 115 bits (288), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 54/96 (56%), Positives = 69/96 (71%), Gaps = 0/96 (0%)
Query 20 SASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTT 79
S S + GL+ +Y + L PLE ++F F SPLLT+ DF AKPMVM+LGQYSTGKTT
Sbjct 93 SLSSVTSIVDGLKRLYIQKLKPLEVAYRFNDFVSPLLTNSDFDAKPMVMLLGQYSTGKTT 152
Query 80 FVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
F++HLL+ YPG IGPEPTTD+FV +M G ++ I
Sbjct 153 FIKHLLKSTYPGAHIGPEPTTDRFVVVMSGPDERSI 188
> HsM7657054
Length=543
Score = 115 bits (287), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 72/111 (64%), Gaps = 2/111 (1%)
Query 5 MSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAK 64
M WL R Q V + L+ +Y+ LLPLE ++F F+SP L D DF K
Sbjct 1 MFSWL--KRGGARGQQPEAIRTVTSALKELYRTKLLPLEEHYRFGAFHSPALEDADFDGK 58
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
PMV+V GQYSTGKT+F+Q+LLE++ PG R+GPEPTTD FVA+M G T+ +
Sbjct 59 PMVLVAGQYSTGKTSFIQYLLEQEVPGSRVGPEPTTDFFVAVMHGDTEGTV 109
> Hs7657056
Length=535
Score = 108 bits (269), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 53/98 (54%), Positives = 76/98 (77%), Gaps = 0/98 (0%)
Query 18 QQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGK 77
++ V+ V GL+ +YK LLPLE ++F +F+SP L D DF KPMV+++GQYSTGK
Sbjct 12 RKDPEVFQTVSEGLKKLYKSKLLPLEEHYRFHEFHSPALEDADFDNKPMVLLVGQYSTGK 71
Query 78 TTFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
TTF+++LLE+D+PG+RIGPEPTTD F+A+M+G + +I
Sbjct 72 TTFIRYLLEQDFPGMRIGPEPTTDSFIAVMQGDMEGII 109
> At4g05520
Length=514
Score = 105 bits (263), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 52/97 (53%), Positives = 64/97 (65%), Gaps = 11/97 (11%)
Query 30 GLQAMYKKYLLPLESDFKFQQFYSPLL-----------TDGDFAAKPMVMVLGQYSTGKT 78
GL+ +Y + L PLE ++F F SP+L T DF AKPMVM+LGQYSTGKT
Sbjct 169 GLKRLYTEKLKPLEVTYRFNDFASPVLVSNILTVSETLTSSDFDAKPMVMLLGQYSTGKT 228
Query 79 TFVQHLLERDYPGLRIGPEPTTDKFVAIMEGSTDQVI 115
TF++HLL DYPG IGPEPTTD+FV M G ++ I
Sbjct 229 TFIKHLLGCDYPGAHIGPEPTTDRFVVAMSGPDERTI 265
> Hs18586357
Length=355
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 50/88 (56%), Gaps = 2/88 (2%)
Query 24 YNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQH 83
Y+ V L+ +Y + PLE +K+ + +TDG+ +KPMV+ LG +S GK+T + +
Sbjct 11 YSAVLQRLRKIYHSSIKPLEQSYKYNELRQHEITDGEITSKPMVLFLGPWSVGKSTMINY 70
Query 84 L--LERDYPGLRIGPEPTTDKFVAIMEG 109
L LE L G EPTT +F +M G
Sbjct 71 LLGLENTRYQLYTGAEPTTSEFTVLMHG 98
> 7299809_2
Length=417
Score = 50.8 bits (120), Expect = 7e-07, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 31 LQAMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDYP 90
++ +Y+ + PLE+ +K++ + +D + +KP+++ +G +S GK++ + +L + +Y
Sbjct 4 IKRIYENAVKPLETLYKYRDLSNRHFSDPEIFSKPLILFMGPWSGGKSSILNYLTDNEYT 63
Query 91 --GLRIGPEPTTDKFVAIMEGSTDQVI 115
LR G EP+ F +M G+ +V+
Sbjct 64 PNSLRTGAEPSPAYFNILMWGNETEVL 90
> At4g19210
Length=600
Score = 33.9 bits (76), Expect = 0.078, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 40/77 (51%), Gaps = 10/77 (12%)
Query 35 YKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHL------LERD 88
Y +Y P + K Q + +++G+F ++++LG+ TGKTTF++ L + R+
Sbjct 347 YARYKYPTMT--KTQGNFRLRVSEGEFTDSQIIVMLGENGTGKTTFIRMLVFSSSMIYRE 404
Query 89 YPGLRIG--PEPTTDKF 103
P + P+ + KF
Sbjct 405 IPEFNVSYKPQKISPKF 421
> SPAC4F10.11
Length=469
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 24/42 (57%), Gaps = 9/42 (21%)
Query 67 VMVLGQYSTGKTTFVQHLLERD-YPGLR--------IGPEPT 99
V+VLG+ +GK+T V LL RD YP + + PEPT
Sbjct 98 VLVLGESGSGKSTLVNTLLNRDVYPPTQKSLTGDFGVNPEPT 139
> CE01182
Length=355
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 22/43 (51%), Gaps = 4/43 (9%)
Query 64 KPMVMVLGQYSTGKTTFVQHLL----ERDYPGLRIGPEPTTDK 102
KP ++VLG +GKTTFVQ L R P I +P K
Sbjct 30 KPSILVLGMAGSGKTTFVQRLTAFLHARKTPPYVINLDPAVSK 72
> At4g30300
Length=181
Score = 31.6 bits (70), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 31/58 (53%), Gaps = 10/58 (17%)
Query 45 DFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHL----LERDYPGLRIGPEP 98
DFK + L G+F ++++LG+ TGKTTF++ L L+ D G ++ P
Sbjct 7 DFKLR------LNQGEFTDSQIIVMLGENGTGKTTFIKMLAGSKLDIDEEGSQVHEIP 58
> Hs5031795
Length=352
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 51 FYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDYPG---LRIGPE 97
F+ L ++ + K V+VLG ++T T Q+LLE D G L +GPE
Sbjct 136 FFVQLPSNTTWTLKVRVLVLGPHNTNLTLATQYLLEVDASGWHQLPLGPE 185
> At3g13640
Length=603
Score = 31.6 bits (70), Expect = 0.42, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 10/54 (18%)
Query 35 YKKYLLPLES----DFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHL 84
Y +Y P + DFK + + +G+F ++++LG+ TGKTTF++ L
Sbjct 347 YARYKYPNMTKQLGDFKLE------VMEGEFTDSQIIVMLGENGTGKTTFIRML 394
> SPBC14F5.06
Length=593
Score = 30.8 bits (68), Expect = 0.64, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 43 ESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPTTDK 102
+ DFK + G F+ ++++LG+ TGKTTF + + + + + P+ K
Sbjct 360 QGDFKLT------IKSGGFSDAEIIVLLGENGTGKTTFCKWMAKNSDLKISMKPQTIAPK 413
Query 103 F 103
F
Sbjct 414 F 414
> Hs4506559
Length=599
Score = 30.0 bits (66), Expect = 0.95, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 59 GDFAAKPMVMVLGQYSTGKTTFVQHLLERDYP 90
G+F ++++LG+ TGKTTF++ L R P
Sbjct 367 GEFTDSEIMVMLGENGTGKTTFIRMLAGRLKP 398
> Hs20532766
Length=599
Score = 30.0 bits (66), Expect = 0.98, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 59 GDFAAKPMVMVLGQYSTGKTTFVQHLLERDYP 90
G+F ++++LG+ TGKTTF++ L R P
Sbjct 367 GEFTDSEIMVMLGENGTGKTTFIRMLAGRLKP 398
> CE29474
Length=700
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 47 KFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDY 89
K Q ++ L D P ++V+G S GK++ +++L+ RD+
Sbjct 10 KLQDVFATLGRKEDQIQLPQIVVVGSQSAGKSSVLENLVGRDF 52
> At1g03160
Length=642
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 28/40 (70%), Gaps = 3/40 (7%)
Query 64 KPMVMVL-GQYSTGKTTFVQHLLERDYPGLRIGPEPTTDK 102
+P +MV+ G++++GK+T + LL + Y L+ G PTT++
Sbjct 348 EPFLMVIVGEFNSGKSTVINALLGKRY--LKEGVVPTTNE 385
> 7295014
Length=611
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 29/54 (53%), Gaps = 9/54 (16%)
Query 59 GDFAAKPMVMVLGQYSTGKTTFVQHLL-------ERDYPGLRIG--PEPTTDKF 103
G F+ ++++LG+ TGKTTF++ L E + P L I P+ + KF
Sbjct 375 GHFSDSEILVLLGENGTGKTTFIRMLAGNLQPDGEVELPMLNISYKPQKISPKF 428
> Hs7706300
Length=350
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query 58 DGDFAAKPMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPT 99
DG A+ V +G + GKTT + L+RD P P+PT
Sbjct 25 DGAEIAEKFVFFIGSKNGGKTTIILRCLDRDEP-----PKPT 61
> Hs14727149
Length=351
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query 58 DGDFAAKPMVMVLGQYSTGKTTFVQHLLERDYPGLRIGPEPT 99
DG A+ V +G + GKTT + L+RD P P+PT
Sbjct 25 DGAEIAEKFVFFIGSKNGGKTTIILRCLDRDEP-----PKPT 61
> At1g80480
Length=444
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 65 PMVMVLGQYSTGKTTFVQHLLERDYPGLRIG 95
P ++ G +GKTT + H+L RD+ G RI
Sbjct 90 PATIITGFLGSGKTTLLNHILTRDH-GKRIA 119
> At4g33650
Length=808
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 47 KFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDY 89
K Q ++ L + A P V+V+G S+GK++ ++ L+ RD+
Sbjct 43 KLQDIFAQLGSQSTIAL-PQVVVVGSQSSGKSSVLEALVGRDF 84
> ECU09g1800
Length=934
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 39/86 (45%), Gaps = 7/86 (8%)
Query 1 AGGKMSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLPLESDFKFQQFYSPLLTDGD 60
AGG + F + + T S+SV ++ K + +YK+ L L D + F S
Sbjct 582 AGGDIGGTTFSNLEGMTSCSSSVLDEHKVSIPKVYKEEFLRLNDDQRSALFLS------- 634
Query 61 FAAKPMVMVLGQYSTGKTTFVQHLLE 86
+ ++ G TGK+T + L++
Sbjct 635 LNCRNYRIIHGMPGTGKSTLICLLIK 660
> CE21713
Length=610
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 10/26 (38%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 59 GDFAAKPMVMVLGQYSTGKTTFVQHL 84
GDF+ ++++LG+ TGKTT ++ +
Sbjct 376 GDFSDSEIIVMLGENGTGKTTMIKMM 401
> At1g15730
Length=448
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Query 14 KSGTQQSASVYNDVKAGLQ-AMYKKYLLPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQ 72
KS + SA++Y+D + A+ L + D S +L D P ++ G
Sbjct 44 KSSSFYSAALYSDSRRRFHSAVASDSSLAVVDDEDIFDVASEILPDNRI---PATIITGF 100
Query 73 YSTGKTTFVQHLLERDYPGLRIG 95
+GKTT + H+L D+ G RI
Sbjct 101 LGSGKTTLLNHILTGDH-GKRIA 122
> CE21915
Length=169
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 3 GKMSRWLFGSRKSGTQQSASVYNDVKAGLQAMYKKYLLP 41
G++ ++++ S S + VYN +K + MYK L P
Sbjct 118 GELEKFIYSSNISDGGNATCVYNAIKNEARFMYKNELKP 156
> At2g14120
Length=782
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 47 KFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLERDY 89
K Q ++ L + A P V V+G S+GK++ ++ L+ RD+
Sbjct 27 KLQDIFAQLGSQSTIAL-PQVAVVGSQSSGKSSVLEALVGRDF 68
> Hs18594546_1
Length=337
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 40/100 (40%), Gaps = 23/100 (23%)
Query 8 WLFGSRKS-------GTQQSASVYNDVKAGLQAMYK---------------KYLLPLESD 45
WL G K+ G QQ A V + AG Q+ ++ E +
Sbjct 230 WLPGDHKAPGRHQAIGEQQDA-VDHQATAGYQSPVNTRPQWDTDLRQTSSPRWTYRPEVE 288
Query 46 FKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLL 85
F + + P L+ FA + M +VLG S +T QHL+
Sbjct 289 FTLRGTFGPSLSKWKFAFRQMSVVLGHTSLMSSTTSQHLV 328
> CE08535
Length=1927
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 47 KFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLLER 87
+ ++SPL + F ++M +G T T F+ HLL R
Sbjct 711 QIHNYFSPLCRNEMFDRDVLMMQVGAALTPSTKFIFHLLHR 751
> 7290105
Length=382
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 56 LTDGDFAAKPMVMVLGQYSTGKTTFVQHLLER 87
L++G + ++VLG +GKTTF Q L++
Sbjct 15 LSEGIRESPVCILVLGMAGSGKTTFTQKLIQH 46
> YDR091c
Length=608
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 27/48 (56%), Gaps = 7/48 (14%)
Query 58 DGDFAAKPMVMVLGQYSTGKTTFVQHLL-------ERDYPGLRIGPEP 98
+G+F+ +++++G+ TGKTT ++ L +D P L + +P
Sbjct 372 EGEFSDSEILVMMGENGTGKTTLIKLLAGALKPDEGQDIPKLNVSMKP 419
> Hs16166977
Length=124
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 60 DFAAKPMVMVLGQYSTGKTTFVQHLLERDYP 90
+F ++++LG+ TGKTTF++ L R P
Sbjct 24 EFTDSEIMVMLGENGTGKTTFIRMLAGRLKP 54
> CE24831
Length=389
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 26/64 (40%), Gaps = 2/64 (3%)
Query 28 KAGLQAMYKKYL--LPLESDFKFQQFYSPLLTDGDFAAKPMVMVLGQYSTGKTTFVQHLL 85
K Q K YL P++S QF+S L DG F M + LG T +
Sbjct 305 KTKAQKTIKNYLSTAPIKSTISDDQFFSALGEDGLFIMDQMALNLGDIPASYLTISMRNI 364
Query 86 ERDY 89
+D+
Sbjct 365 CQDF 368
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1178471386
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40