bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6548_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
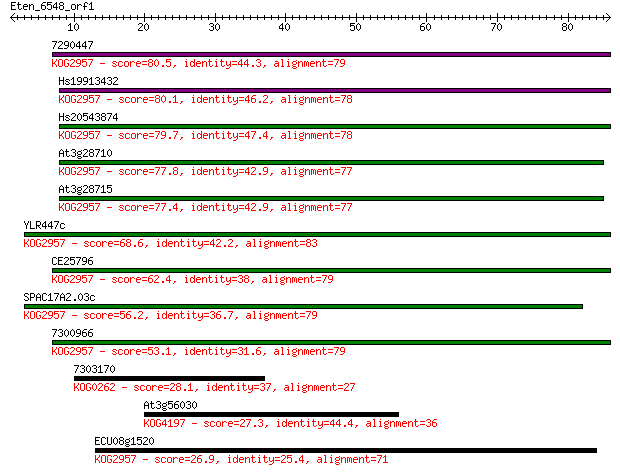
7290447 80.5 8e-16
Hs19913432 80.1 8e-16
Hs20543874 79.7 1e-15
At3g28710 77.8 4e-15
At3g28715 77.4 7e-15
YLR447c 68.6 3e-12
CE25796 62.4 2e-10
SPAC17A2.03c 56.2 1e-08
7300966 53.1 1e-07
7303170 28.1 3.9
At3g56030 27.3 7.7
ECU08g1520 26.9 9.4
> 7290447
Length=350
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 52/79 (65%), Gaps = 0/79 (0%)
Query 7 LFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAV 66
+FN+ +GYLEG+ RG + L + DY L +TLEDL+ L+ TDYG F+ +EP PL+V
Sbjct 7 MFNIDNGYLEGLCRGFKCGILKQADYLNLVQCETLEDLKLHLQGTDYGSFLANEPSPLSV 66
Query 67 PTIAQRARSKLTAEFKYLQ 85
I + R KL EF++++
Sbjct 67 SVIDDKLREKLVIEFQHMR 85
> Hs19913432
Length=351
Score = 80.1 bits (196), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 36/78 (46%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV +GYLEG+VRGL+ L++ DY L +TLEDL+ L+ TDYG F+ +E PL V
Sbjct 9 FNVDNGYLEGLVRGLKAGVLSQADYLNLVQCETLEDLKLHLQSTDYGNFLANEASPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I R + K+ EF++++
Sbjct 69 VIDDRLKEKMVVEFRHMR 86
> Hs20543874
Length=350
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV GYLEG+VRG + S LT++DY L +TLEDL+ L+ TDYG F+ + PL V
Sbjct 9 FNVDHGYLEGLVRGCKASLLTQQDYINLVQCETLEDLKIHLQTTDYGNFLANHTNPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I R +L EF+Y +
Sbjct 69 KIDTEMRKRLCGEFEYFR 86
> At3g28710
Length=351
Score = 77.8 bits (190), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FN+H GYLE IVRG R LT DY L + L+D++ L T YG ++Q+EP PL
Sbjct 9 FNIHGGYLEAIVRGHRAGLLTTADYNNLCQCENLDDIKMHLSATKYGSYLQNEPSPLHTT 68
Query 68 TIAQRARSKLTAEFKYL 84
TI ++ KL ++K++
Sbjct 69 TIVEKCTLKLVDDYKHM 85
> At3g28715
Length=351
Score = 77.4 bits (189), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FN+H GYLE IVRG R LT DY L + L+D++ L T YG ++Q+EP PL
Sbjct 9 FNIHGGYLEAIVRGHRAGLLTTADYNNLCQCENLDDIKMHLSATKYGPYLQNEPSPLHTT 68
Query 68 TIAQRARSKLTAEFKYL 84
TI ++ KL ++K++
Sbjct 69 TIVEKCTLKLVDDYKHM 85
> YLR447c
Length=345
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQD-EP 61
ME FN+ +G++EG+VRG R L+ Y L DTLEDL+ L TDYG F+
Sbjct 1 MEGVYFNIDNGFIEGVVRGYRNGLLSNNQYINLTQCDTLEDLKLQLSSTDYGNFLSSVSS 60
Query 62 LPLAVPTIAQRARSKLTAEFKYLQ 85
L I + A SKL EF Y++
Sbjct 61 ESLTTSLIQEYASSKLYHEFNYIR 84
> CE25796
Length=343
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 48/79 (60%), Gaps = 5/79 (6%)
Query 7 LFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAV 66
LFN+ GYLE + RGL+G L + DY A+ ++ L+ ++ TDYG F+ +EP + V
Sbjct 5 LFNIDHGYLEALTRGLKGGLLAQADY-----ANLVQYLKLHIQSTDYGNFLANEPGAITV 59
Query 67 PTIAQRARSKLTAEFKYLQ 85
I ++ + KL EF +L+
Sbjct 60 QVIDEKLKEKLVTEFTHLR 78
> SPAC17A2.03c
Length=343
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 1/79 (1%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPL 62
M+ FN + GY+E +VRG + L + Y L+ ++LED R L TDYG F+ ++
Sbjct 1 MDALSFNTNSGYIEALVRGYESALLEQHIYSNLSQCESLEDFRLQLSSTDYGGFLANQS- 59
Query 63 PLAVPTIAQRARSKLTAEF 81
L I+ +A KL EF
Sbjct 60 KLTSSIISAKATEKLLDEF 78
> 7300966
Length=350
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 41/79 (51%), Gaps = 0/79 (0%)
Query 7 LFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAV 66
+FN GYLE + RG + L DY L ++LED+ +++ TDYG E +V
Sbjct 4 IFNTEYGYLEALTRGFKNGMLKHSDYLNLTQCESLEDVMISIQGTDYGLIFGGEQSAPSV 63
Query 67 PTIAQRARSKLTAEFKYLQ 85
I + R +L ++ Y++
Sbjct 64 EVIERCLRDRLLQQYYYIR 82
> 7303170
Length=1642
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 10 VHDGYLEGIVRGLRGSFLTREDYKRLA 36
+ D + G+ +RG F REDY++L
Sbjct 608 IQDHVISGVKLSIRGRFFNREDYQQLV 634
> At3g56030
Length=336
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query 20 RGLRGSFLTREDYKRLANADT----LEDLRSALEETDYGF 55
R + F T +K L N D+ LEDLR L +TD G+
Sbjct 68 RVVNACFNTTATFKFLTNTDSYSSSLEDLRRILPQTDAGY 107
> ECU08g1520
Length=341
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 31/71 (43%), Gaps = 3/71 (4%)
Query 13 GYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVPTIAQR 72
GY+ + G + L EDY L + LE++ L +T + E + P + +R
Sbjct 24 GYIISEINGKKEEMLKEEDYNALKRCENLEEVAIKLSKT---YRSLSEGIAYTKPELKKR 80
Query 73 ARSKLTAEFKY 83
L A+F +
Sbjct 81 LLETLKADFDH 91
Lambda K H
0.320 0.138 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194132014
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40