bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6530_orf2
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
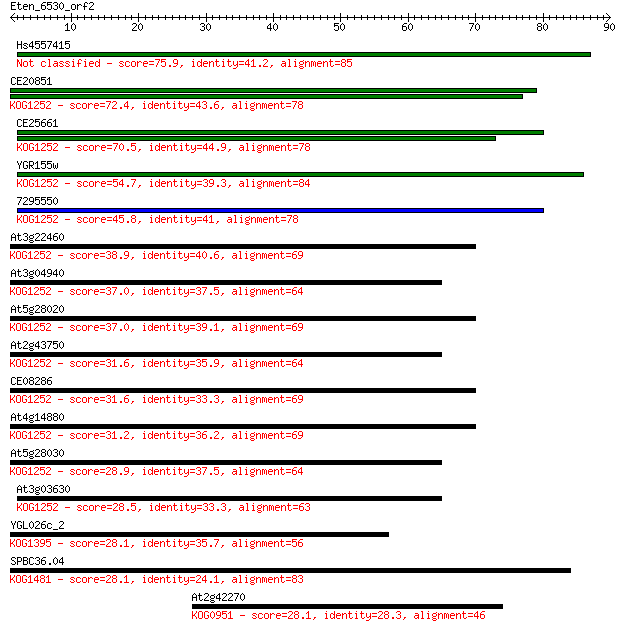
Hs4557415 75.9 2e-14
CE20851 72.4 2e-13
CE25661 70.5 7e-13
YGR155w 54.7 5e-08
7295550 45.8 2e-05
At3g22460 38.9 0.002
At3g04940 37.0 0.009
At5g28020 37.0 0.010
At2g43750 31.6 0.40
CE08286 31.6 0.41
At4g14880 31.2 0.45
At5g28030 28.9 2.4
At3g03630 28.5 3.3
YGL026c_2 28.1 3.8
SPBC36.04 28.1 4.3
At2g42270 28.1 4.4
> Hs4557415
Length=551
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/85 (41%), Positives = 55/85 (64%), Gaps = 4/85 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND E+ +R+LI EG+L GGS+G+ +A A+KA + + +R ++LPD RN
Sbjct 325 KSNDEEAFTFARMLIAQEGLLCGGSAGSTVAVAVKAAQEL----QEGQRCVVILPDSVRN 380
Query 62 YTSKFVCDEWMAEKGFLERSELSRQ 86
Y +KF+ D WM +KGFL+ +L+ +
Sbjct 381 YMTKFLSDRWMLQKGFLKEEDLTEK 405
> CE20851
Length=755
Score = 72.4 bits (176), Expect = 2e-13, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 53/78 (67%), Gaps = 2/78 (2%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+K +D ES L +R +IR EG+L GGSSG A+ AL+ ++ +D++ + ++LPDG R
Sbjct 587 LKSDDKESFLMAREIIRTEGILCGGSSGCAVHYALEQCRKLDLPEDAN--VVVLLPDGIR 644
Query 61 NYTSKFVCDEWMAEKGFL 78
NY +KF+ D+WM +GF
Sbjct 645 NYLTKFLDDDWMKARGFF 662
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 9/77 (11%)
Query 1 VKVNDA-ESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGS 59
V++ DA ES +R LI EG++ G SSGAA+ A+K + + + + +VL DG
Sbjct 229 VEIRDAPESYTFTRHLIETEGIMAGPSSGAAVLEAIKLAKDL----PAGSVVVVVLMDGI 284
Query 60 RNYTSKFVCDEWMAEKG 76
R+Y +WM G
Sbjct 285 RDYLDA----DWMKVNG 297
> CE25661
Length=700
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 53/78 (67%), Gaps = 2/78 (2%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K +D ES L +R LIR EG+L GGSSG A+ AL+ + + +++ + ++LPDG RN
Sbjct 624 KSHDKESFLMARELIRSEGILCGGSSGCAVHYALEECKSLNLPAEAN--VVVLLPDGIRN 681
Query 62 YTSKFVCDEWMAEKGFLE 79
Y +KF+ D+WM E+ FL+
Sbjct 682 YITKFLDDDWMNERHFLD 699
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 4/71 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
+V + E+ +R LI EG++ G SSGAA+ A+K + + + I +VL DG RN
Sbjct 246 EVEEDEAYSFTRHLIGTEGIMAGPSSGAAVLEAIKLAKEL----PAGSTIVVVLMDGIRN 301
Query 62 YTSKFVCDEWM 72
Y F+ D+W+
Sbjct 302 YLRHFLDDDWI 312
> YGR155w
Length=507
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 33/86 (38%), Positives = 46/86 (53%), Gaps = 5/86 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMG--WKDDSSKRIAMVLPDGS 59
K +D S +R LI EG+LVGGSSG+A +K E +DD I + PD
Sbjct 265 KTDDKPSFKYARQLISNEGVLVGGSSGSAFTAVVKYCEDHPELTEDDV---IVAIFPDSI 321
Query 60 RNYTSKFVCDEWMAEKGFLERSELSR 85
R+Y +KFV DEW+ + + L+R
Sbjct 322 RSYLTKFVDDEWLKKNNLWDDDVLAR 347
> 7295550
Length=522
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 45/78 (57%), Gaps = 4/78 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K+ D++ SR L EG+L GGSSG A+ AL+ ++ +R ++LPDG RN
Sbjct 294 KIGDSDCFPMSRRLNAEEGLLCGGSSGGAMHAALEHARKL----KKGQRCVVILPDGIRN 349
Query 62 YTSKFVCDEWMAEKGFLE 79
Y +KFV D WM + F E
Sbjct 350 YMTKFVSDNWMEARNFKE 367
> At3g22460
Length=321
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 28/69 (40%), Positives = 39/69 (56%), Gaps = 3/69 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
V+V+ ES+ +RLL R EG+LVG SSGAA A+K +R +++ K I V P
Sbjct 243 VQVSSEESIDMARLLAREEGLLVGISSGAAATAAIKLAKR---PENAGKLIVAVFPSFGE 299
Query 61 NYTSKFVCD 69
Y S + D
Sbjct 300 RYLSTVLFD 308
> At3g04940
Length=399
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 38/64 (59%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGAA A A+K +R +++ K I ++ P G
Sbjct 322 IQVTSVEAIETAKLLALKEGLLVGISSGAAAAAAIKVAKR---PENAGKLIVVIFPSGGE 378
Query 61 NYTS 64
Y S
Sbjct 379 RYLS 382
> At5g28020
Length=323
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGAA A ALK +R +++ K I +V P G
Sbjct 246 IQVAGEEAIETAKLLALKEGLLVGISSGAAAAAALKVAKR---PENAGKLIVVVFPSGGE 302
Query 61 NYTSKFVCD 69
Y S + D
Sbjct 303 RYLSTKLFD 311
> At2g43750
Length=392
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+ ++ E++ S+ L EG+LVG SSGAA A A++ +R +++ K IA+V P
Sbjct 314 IAISSEEAIETSKQLALQEGLLVGISSGAAAAAAIQVAKR---PENAGKLIAVVFPSFGE 370
Query 61 NYTS 64
Y S
Sbjct 371 RYLS 374
> CE08286
Length=344
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPD-GS 59
++++ E+++ ++ L EG+L G SSGA +A AL+ R + + K I LP G
Sbjct 249 IRIHSDEAIVMAQRLSYEEGLLGGISSGANVAAALQLAAR---PEMAGKLIVTCLPSCGE 305
Query 60 RNYTSKFVCD 69
R TS D
Sbjct 306 RYMTSPLYTD 315
> At4g14880
Length=322
Score = 31.2 bits (69), Expect = 0.45, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
V+V+ ES+ +R L EG+LVG SSGAA A A+K +R +++ K + P
Sbjct 244 VQVSSDESIDMARQLALKEGLLVGISSGAAAAAAIKLAQR---PENAGKLFVAIFPSFGE 300
Query 61 NYTSKFVCD 69
Y S + D
Sbjct 301 RYLSTVLFD 309
> At5g28030
Length=323
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGA+ A ALK +R ++ K I ++ P G
Sbjct 246 IQVTGEEAIETTKLLAIKEGLLVGISSGASAAAALKVAKR---PENVGKLIVVIFPSGGE 302
Query 61 NYTS 64
Y S
Sbjct 303 RYLS 306
> At3g03630
Length=404
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
KV++ E++ +R L EG+LVG SSGAA A+ +R +++ K I ++ P
Sbjct 329 KVSNGEAIEMARRLALEEGLLVGISSGAAAVAAVSLAKR---AENAGKLITVLFPSHGER 385
Query 62 YTS 64
Y +
Sbjct 386 YIT 388
> YGL026c_2
Length=405
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 11/67 (16%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERM-----------GWKDDSSK 49
+ DA++LL +LL +LEG++ S A+ GA + + M G D +
Sbjct 321 IAATDAQALLGFKLLSQLEGIIPALESSHAVYGACELAKTMKPDQHLVINISGRGDKDVQ 380
Query 50 RIAMVLP 56
+A VLP
Sbjct 381 SVAEVLP 387
> SPBC36.04
Length=351
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 39/83 (46%), Gaps = 10/83 (12%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+K+ D +S+ L+ EG+ +GGSS + A++ + +G K + +L D
Sbjct 264 MKIPDEKSINMFFRLLDQEGLFLGGSSCLNVVAAVEMAKILG----PGKTVVTILCDSGH 319
Query 61 NYTSKFVCDEWMAEKGFLERSEL 83
Y ++ + + FLE +L
Sbjct 320 KYATR------LFSRSFLESKKL 336
> At2g42270
Length=2172
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 28 GAALAGALKAIERMGWKDDSSKRIAMVLPD--GSRNYTSKFVCDEWMA 73
G A L AI+R+ + + ++ +P G ++YT F+CD ++
Sbjct 2103 GEAKTNQLMAIKRISLQRKAQVKLEFAVPTETGEKSYTLYFMCDSYLG 2150
Lambda K H
0.318 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181033294
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40