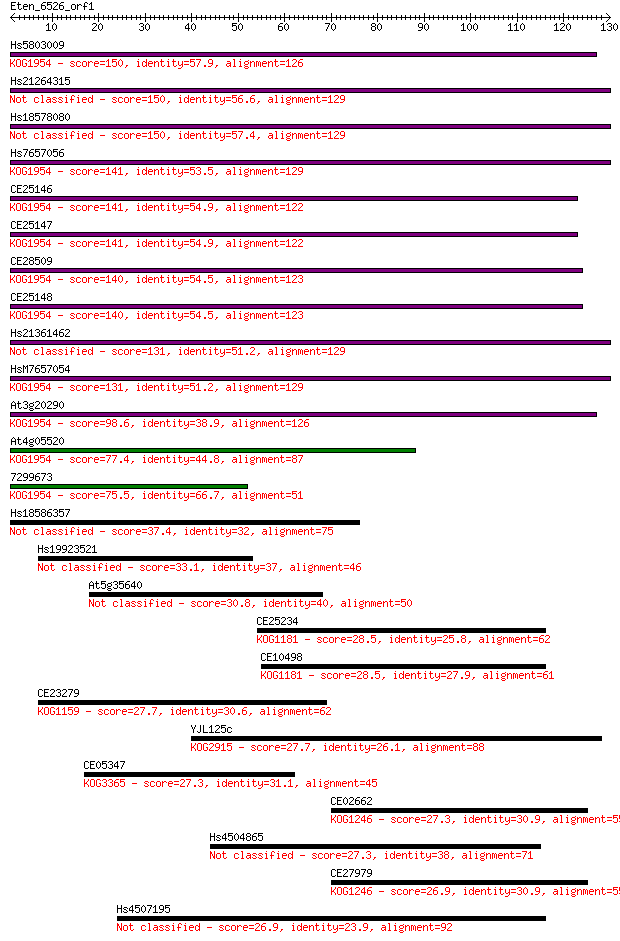
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6526_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
Hs5803009 150 4e-37
Hs21264315 150 4e-37
Hs18578080 150 4e-37
Hs7657056 141 2e-34
CE25146 141 3e-34
CE25147 141 3e-34
CE28509 140 5e-34
CE25148 140 5e-34
Hs21361462 131 2e-31
HsM7657054 131 3e-31
At3g20290 98.6 2e-21
At4g05520 77.4 6e-15
7299673 75.5 2e-14
Hs18586357 37.4 0.007
Hs19923521 33.1 0.12
At5g35640 30.8 0.73
CE25234 28.5 3.0
CE10498 28.5 3.1
CE23279 27.7 5.4
YJL125c 27.7 6.1
CE05347 27.3 6.9
CE02662 27.3 7.9
Hs4504865 27.3 8.1
CE27979 26.9 8.5
Hs4507195 26.9 10.0
> Hs5803009
Length=534
Score = 150 bits (380), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 73/127 (57%), Positives = 99/127 (77%), Gaps = 1/127 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW++PL +NR+LFEAE DL+ DI +PR+AA+RKLND IKRARLAKVH
Sbjct 248 EVVRVYIGSFWSHPLLIPDNRKLFEAEEQDLFKDIQSLPRNAALRKLNDLIKRARLAKVH 307
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++++L+ ++P +FGKESKKK+L+N LG IYQ++ +E+ + GDFP L MQE L + +
Sbjct 308 AYIISSLKKEMPNVFGKESKKKELVNNLGEIYQKIEREHQISPGDFPSLRKMQELLQTQD 367
Query 120 WSKIPRL 126
+SK L
Sbjct 368 FSKFQAL 374
> Hs21264315
Length=541
Score = 150 bits (380), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 73/130 (56%), Positives = 100/130 (76%), Gaps = 1/130 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW PLQN +NRRLFEAEA DL+ DI +P+ AA+RKLND IKRARLAKVH
Sbjct 251 EVLRVYIGSFWAQPLQNTDNRRLFEAEAQDLFRDIQSLPQKAAVRKLNDLIKRARLAKVH 310
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A +++ L+ ++P +FGKE+KK++LI++L IY Q+ +EY + GDFP + MQE+L +++
Sbjct 311 AYIISYLKKEMPSVFGKENKKRELISRLPEIYIQLQREYQISAGDFPEVKAMQEQLENYD 370
Query 120 WSKIPRLDMK 129
++K L K
Sbjct 371 FTKFHSLKPK 380
> Hs18578080
Length=534
Score = 150 bits (380), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 74/130 (56%), Positives = 100/130 (76%), Gaps = 1/130 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW++PL +NR+LFEAE DL+ DI +PR+AA+RKLND IKRARLAKVH
Sbjct 248 EVVRVYIGSFWSHPLLIPDNRKLFEAEEQDLFKDIQSLPRNAALRKLNDLIKRARLAKVH 307
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++++L+ ++P +FGKESKKK+L+N LG IYQ++ +E+ + GDFP L MQE L + +
Sbjct 308 AYIISSLKKEMPNVFGKESKKKELVNNLGEIYQKIEREHQISPGDFPSLRKMQELLQTQD 367
Query 120 WSKIPRLDMK 129
+SK L K
Sbjct 368 FSKFQALKPK 377
> Hs7657056
Length=535
Score = 141 bits (356), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 69/130 (53%), Positives = 99/130 (76%), Gaps = 1/130 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW++PL +NR+LFEAE DL+ DI +PR+AA+RKLND IKRARLAKVH
Sbjct 248 EVIRVYIGSFWSHPLLIPDNRKLFEAEEQDLFRDIQSLPRNAALRKLNDLIKRARLAKVH 307
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++++L+ ++P +FGK++KKK+L+N L IY ++ +E+ + GDFP L MQ++L + +
Sbjct 308 AYIISSLKKEMPSVFGKDNKKKELVNNLAEIYGRIEREHQISPGDFPNLKRMQDQLQAQD 367
Query 120 WSKIPRLDMK 129
+SK L K
Sbjct 368 FSKFQPLKSK 377
> CE25146
Length=786
Score = 141 bits (356), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 93/123 (75%), Gaps = 1/123 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV+RVY+GSFW++PL D NRRLF+ E +DL+ D+ +PR+AA+RKLND IKRARLAKVH
Sbjct 479 EVSRVYLGSFWDHPLHYDLNRRLFQDEQHDLFQDLQALPRNAALRKLNDLIKRARLAKVH 538
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++ LR ++P M GK+ KKK LI L IY+Q+ +E+ + GDFP + M+EKL + +
Sbjct 539 AYIIAELRKQMPSMIGKDKKKKDLIQNLDKIYEQLQREHNISPGDFPDVNKMREKLQTQD 598
Query 120 WSK 122
+SK
Sbjct 599 FSK 601
> CE25147
Length=835
Score = 141 bits (355), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 93/123 (75%), Gaps = 1/123 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV+RVY+GSFW++PL D NRRLF+ E +DL+ D+ +PR+AA+RKLND IKRARLAKVH
Sbjct 528 EVSRVYLGSFWDHPLHYDLNRRLFQDEQHDLFQDLQALPRNAALRKLNDLIKRARLAKVH 587
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++ LR ++P M GK+ KKK LI L IY+Q+ +E+ + GDFP + M+EKL + +
Sbjct 588 AYIIAELRKQMPSMIGKDKKKKDLIQNLDKIYEQLQREHNISPGDFPDVNKMREKLQTQD 647
Query 120 WSK 122
+SK
Sbjct 648 FSK 650
> CE28509
Length=571
Score = 140 bits (353), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 67/124 (54%), Positives = 93/124 (75%), Gaps = 1/124 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV+RVY+GSFW++PL D NRRLF+ E +DL+ D+ +PR+AA+RKLND IKRARLAKVH
Sbjct 264 EVSRVYLGSFWDHPLHYDLNRRLFQDEQHDLFQDLQALPRNAALRKLNDLIKRARLAKVH 323
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++ LR ++P M GK+ KKK LI L IY+Q+ +E+ + GDFP + M+EKL + +
Sbjct 324 AYIIAELRKQMPSMIGKDKKKKDLIQNLDKIYEQLQREHNISPGDFPDVNKMREKLQTQD 383
Query 120 WSKI 123
+SK
Sbjct 384 FSKF 387
> CE25148
Length=589
Score = 140 bits (353), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 67/124 (54%), Positives = 93/124 (75%), Gaps = 1/124 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV+RVY+GSFW++PL D NRRLF+ E +DL+ D+ +PR+AA+RKLND IKRARLAKVH
Sbjct 282 EVSRVYLGSFWDHPLHYDLNRRLFQDEQHDLFQDLQALPRNAALRKLNDLIKRARLAKVH 341
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A ++ LR ++P M GK+ KKK LI L IY+Q+ +E+ + GDFP + M+EKL + +
Sbjct 342 AYIIAELRKQMPSMIGKDKKKKDLIQNLDKIYEQLQREHNISPGDFPDVNKMREKLQTQD 401
Query 120 WSKI 123
+SK
Sbjct 402 FSKF 405
> Hs21361462
Length=543
Score = 131 bits (330), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 66/130 (50%), Positives = 91/130 (70%), Gaps = 1/130 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW+ PL +NRRLFE E DL+ DI +PR AA+RKLND +KRARL +VH
Sbjct 248 EVLRVYIGSFWSQPLLVPDNRRLFELEEQDLFRDIQGLPRHAALRKLNDLVKRARLVRVH 307
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A +++ L+ ++P +FGKE+KKKQLI +L I+ ++ E+ + GDFP MQE L + +
Sbjct 308 AYIISYLKKEMPSVFGKENKKKQLILKLPVIFAKIQLEHHISPGDFPDCQKMQELLMAHD 367
Query 120 WSKIPRLDMK 129
++K L K
Sbjct 368 FTKFHSLKPK 377
> HsM7657054
Length=543
Score = 131 bits (330), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 66/130 (50%), Positives = 91/130 (70%), Gaps = 1/130 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
EV RVYIGSFW+ PL +NRRLFE E DL+ DI +PR AA+RKLND +KRARL +VH
Sbjct 248 EVLRVYIGSFWSQPLLVPDNRRLFELEEQDLFRDIQGLPRHAALRKLNDLVKRARLVRVH 307
Query 61 ALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKLASFE 119
A +++ L+ ++P +FGKE+KKKQLI +L I+ ++ E+ + GDFP MQE L + +
Sbjct 308 AYIISYLKKEMPSVFGKENKKKQLILKLPVIFAKIQLEHHISPGDFPDCQKMQELLMAHD 367
Query 120 WSKIPRLDMK 129
++K L K
Sbjct 368 FTKFHSLKPK 377
> At3g20290
Length=485
Score = 98.6 bits (244), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/131 (37%), Positives = 83/131 (63%), Gaps = 5/131 (3%)
Query 1 EVARVYIGSFWNNPLQNDEN----RRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARL 56
EV+RVYIGSF + P+ R LFE E +DL +D+ +P+ A R++N+F+KRAR
Sbjct 327 EVSRVYIGSFSDKPINEAATGPIGRELFEKEQDDLLADLKDIPKKACDRRINEFVKRARA 386
Query 57 AKVHALLLTTLRNKLP-MFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQEKL 115
AK+HA +++ L+ ++P + GK +++LI+ L + +V +E+ +P GDFP + +E L
Sbjct 387 AKIHAYIISHLKKEMPAIMGKAKAQQKLIDNLEDEFGKVQREHHLPKGDFPNVDHFREVL 446
Query 116 ASFEWSKIPRL 126
+ + K +L
Sbjct 447 SGYNIDKFEKL 457
> At4g05520
Length=514
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 61/92 (66%), Gaps = 5/92 (5%)
Query 1 EVARVYIGSFWNNPLQNDE----NRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARL 56
EV RVYIGSF + P+ + LFE E NDL +D+ VP+ A RK+N+F+KRAR
Sbjct 409 EVVRVYIGSFNDKPINEVAVGPIGKELFEKEQNDLLADLMDVPKKACDRKINEFVKRARS 468
Query 57 AKVHALLLTTLRNKLP-MFGKESKKKQLINQL 87
AK++A +++ L+ ++P M GK +++L++ L
Sbjct 469 AKINAYIMSHLKKEMPAMMGKSKAQQRLMDNL 500
> 7299673
Length=493
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/51 (66%), Positives = 41/51 (80%), Gaps = 0/51 (0%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFI 51
EVARVYIGSFW+ PL+ D NRRLFE E DL+ D+ +PR+AA+RKLND I
Sbjct 245 EVARVYIGSFWDQPLRFDANRRLFEDEEQDLFRDLQSLPRNAALRKLNDLI 295
> Hs18586357
Length=355
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 4/79 (5%)
Query 1 EVARVYIGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVH 60
E RVY+ SFW + D ++ LF E L D+ +V + K+ + A ++H
Sbjct 242 EPPRVYVSSFWPQEYKPDTHQELFLQEEISLLEDLNQVIENRLENKIAFIRQHAIRVRIH 301
Query 61 ALL----LTTLRNKLPMFG 75
ALL L T ++K+ F
Sbjct 302 ALLVDRYLQTYKDKMTFFS 320
> Hs19923521
Length=682
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 7 IGSFWNNPLQNDENRRLFEAEANDLYSDIAR-----VPRDAAIRKLNDFIK 52
+GSFW + DE +RLF + N+ +S+ P++ + LND IK
Sbjct 402 VGSFWTTMAEEDE-QRLFTEKVNNTFSECLNLINEGCPKEDILVTLNDLIK 451
> At5g35640
Length=230
Score = 30.8 bits (68), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 4/54 (7%)
Query 18 DENRRLFEAEANDLYSDIARVP--RDAAIRKLNDFIKRA--RLAKVHALLLTTL 67
D NR A A ++ DI R P D +R+++ FIKRA +L AL T +
Sbjct 23 DINRCSAGAAATSVWWDINRCPLPNDVDVRRVSPFIKRALEKLGYTGALTTTAI 76
> CE25234
Length=4900
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 54 ARLAKVHALLLTTLRNKLPMFGKESKKKQLINQLGTIYQQVSQEYGVPIGDFPPLATMQE 113
AR+ ++ ++ ++TL + S ++ ++ + I + S EY VP+ F P QE
Sbjct 3415 ARITEIASMDISTLPPPTGKPSETSLTQEELDHIARIAEMASAEYDVPVKIFEPPELTQE 3474
Query 114 KL 115
+L
Sbjct 3475 EL 3476
> CE10498
Length=2361
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 55 RLAKVHALLLTTLRNKLPMFGKESKK---KQLINQLGTIYQQVSQEYGVPIGDFPPLATM 111
+AK+ + + P GK S+ ++ ++ + I + S EY VP+ F P
Sbjct 731 HIAKITGMASMDISTLPPPTGKPSETSLTQEELDHIARIAEMASAEYDVPVKIFEPPELT 790
Query 112 QEKL 115
QE+L
Sbjct 791 QEEL 794
> CE23279
Length=585
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 7 IGSFWNNPLQNDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVHAL--LL 64
+G + NP + + + L E D Y D A R L DF+ ++ K L +L
Sbjct 323 LGYYSTNPPEKERLQELASPEGLDDYLDYANRSRRTTAEALRDFVATSKNLKPDYLFEIL 382
Query 65 TTLR 68
TT+R
Sbjct 383 TTIR 386
> YJL125c
Length=383
Score = 27.7 bits (60), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 46/96 (47%), Gaps = 19/96 (19%)
Query 40 RDAAIRKLNDFIKRARLAKVHALLLTTLRNKLPMFGKESKKKQLINQLGTIYQQVSQ--E 97
R +R LND ++R R K H L G E +K+ N + + ++V + E
Sbjct 264 RRQMVRSLNDALERLRDIKRHKLQ-----------GVERRKRMFNNTIDSNDEKVGKRNE 312
Query 98 YGVPIGD---FPPLAT---MQEKLASFEWSKIPRLD 127
GVP+ + F P ++E ++++W ++ +++
Sbjct 313 DGVPLTEKAKFNPFGKGSRIKEGDSNYKWKEVTKME 348
> CE05347
Length=150
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 17 NDENRRLFEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVHA 61
N+ R + + ++PRDAA RK + + + RLA V A
Sbjct 51 NEHPHRALTVVYGRILRALEQIPRDAAYRKYTEAVVKQRLALVQA 95
> CE02662
Length=972
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Query 70 KLPMFGKESKKKQLINQLGTIYQQVS--QEYGVPIGDFPPLATMQEKLASFEWSKIP 124
KLP F +++ L+N G ++ Q Y PIG P +AS W++ P
Sbjct 729 KLPTFLLPNREGNLLNYAGVDVLGINTVQMYAKPIGSRTPAHMENSLMASINWNRGP 785
> Hs4504865
Length=711
Score = 27.3 bits (59), Expect = 8.1, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 40/82 (48%), Gaps = 13/82 (15%)
Query 44 IRK--LNDFIKRAR--LAKVHALLLTTLRNKLPMFGKESKKKQLIN-------QLGTIYQ 92
IRK D ++RAR AK+ TT+ N P FG +K+QL + +L +
Sbjct 69 IRKDAFADAVQRARQIAAKIGGDAATTVNNSTPDFGFGGQKRQLEDGDQPESKKLASQGD 128
Query 93 QVSQEYGVPIGDFPPLATMQEK 114
+S + G PI PP +M E+
Sbjct 129 SISSQLG-PIHP-PPRTSMTEE 148
> CE27979
Length=1013
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Query 70 KLPMFGKESKKKQLINQLGTIYQQVS--QEYGVPIGDFPPLATMQEKLASFEWSKIP 124
KLP F +++ L+N G ++ Q Y PIG P +AS W++ P
Sbjct 770 KLPTFLLPNREGNLLNYAGVDVLGINTVQMYAKPIGSRTPAHMENSLMASINWNRGP 826
> Hs4507195
Length=2364
Score = 26.9 bits (58), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 22/93 (23%), Positives = 42/93 (45%), Gaps = 1/93 (1%)
Query 24 FEAEANDLYSDIARVPRDAAIRKLNDFIKRARLAKVHALLLTTLRNKLPMFGKESKKKQL 83
EA+ +DL + ++ + + + A ++ V + TTL+N+ G+ SK +Q
Sbjct 1008 IEAKLSDLQKEAEKLESEHPDQAQAILSRLAEISDVWEEMKTTLKNREASLGEASKLQQF 1067
Query 84 INQLGTIYQQVSQ-EYGVPIGDFPPLATMQEKL 115
+ L +S+ + + D P T EKL
Sbjct 1068 LRDLDDFQSWLSRTQTAIASEDMPNTLTEAEKL 1100
Lambda K H
0.320 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40