bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6473_orf1
Length=113
Score E
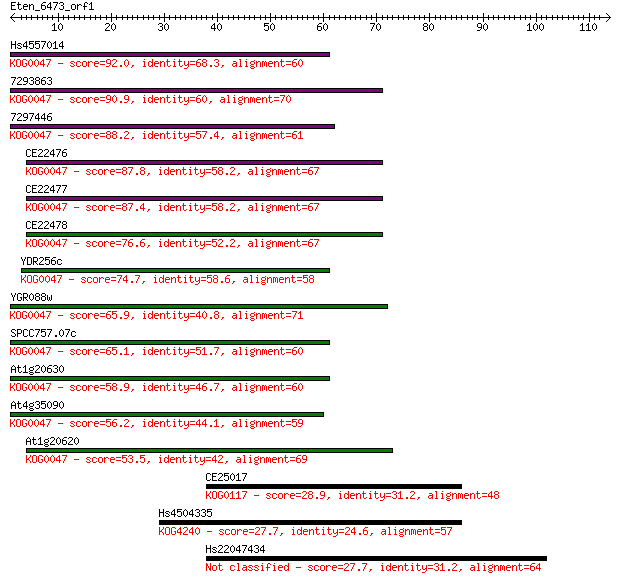
Sequences producing significant alignments: (Bits) Value
Hs4557014 92.0 3e-19
7293863 90.9 5e-19
7297446 88.2 4e-18
CE22476 87.8 5e-18
CE22477 87.4 5e-18
CE22478 76.6 9e-15
YDR256c 74.7 4e-14
YGR088w 65.9 2e-11
SPCC757.07c 65.1 3e-11
At1g20630 58.9 2e-09
At4g35090 56.2 1e-08
At1g20620 53.5 9e-08
CE25017 28.9 2.5
Hs4504335 27.7 5.7
Hs22047434 27.7 6.0
> Hs4557014
Length=527
Score = 92.0 bits (227), Expect = 3e-19, Method: Composition-based stats.
Identities = 41/60 (68%), Positives = 47/60 (78%), Gaps = 0/60 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 60
SL PE+LHQ + LFSDRG PDG+RHM+GY SH FKLVN+ G Y KFH KT+Q IKNLS
Sbjct 187 SLRPESLHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVNANGEAVYCKFHYKTDQGIKNLS 246
> 7293863
Length=506
Score = 90.9 bits (224), Expect = 5e-19, Method: Composition-based stats.
Identities = 42/70 (60%), Positives = 51/70 (72%), Gaps = 0/70 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 60
+L PE+ HQ ILFSDRGTPDGY HM+GY SH FKL+N++G YAKFH KT+Q IKNL
Sbjct 185 TLRPESAHQVCILFSDRGTPDGYCHMNGYGSHTFKLINAKGEPIYAKFHFKTDQGIKNLD 244
Query 61 AAFVFYLSSS 70
L+S+
Sbjct 245 VKTADQLAST 254
> 7297446
Length=506
Score = 88.2 bits (217), Expect = 4e-18, Method: Composition-based stats.
Identities = 35/61 (57%), Positives = 45/61 (73%), Gaps = 0/61 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 60
+L PETLH + FSDRGTPDGYRH+HGY H ++++N+ G Y +FH KT+Q IKNL
Sbjct 185 TLRPETLHALLMYFSDRGTPDGYRHLHGYGVHTYRMINASGETQYVRFHFKTDQGIKNLD 244
Query 61 A 61
A
Sbjct 245 A 245
> CE22476
Length=500
Score = 87.8 bits (216), Expect = 5e-18, Method: Composition-based stats.
Identities = 39/67 (58%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 4 PETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSAAF 63
PE LHQ LFSDRG PDGYRHM+GY SH FK+VN +G Y KFH K Q +KNL+
Sbjct 186 PEALHQVMFLFSDRGLPDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEK 245
Query 64 VFYLSSS 70
L+SS
Sbjct 246 AGQLASS 252
> CE22477
Length=524
Score = 87.4 bits (215), Expect = 5e-18, Method: Composition-based stats.
Identities = 39/67 (58%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 4 PETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSAAF 63
PE LHQ LFSDRG PDGYRHM+GY SH FK+VN +G Y KFH K Q +KNL+
Sbjct 210 PEALHQVMFLFSDRGLPDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEK 269
Query 64 VFYLSSS 70
L+SS
Sbjct 270 AGQLASS 276
> CE22478
Length=512
Score = 76.6 bits (187), Expect = 9e-15, Method: Composition-based stats.
Identities = 35/67 (52%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 4 PETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSAAF 63
PE++HQ L+SDRG PDG+R M+GY +H FK+VN EG Y KFH K Q KNL
Sbjct 201 PESIHQVMFLYSDRGIPDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTD 260
Query 64 VFYLSSS 70
L+SS
Sbjct 261 AGKLASS 267
> YDR256c
Length=515
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 34/61 (55%), Positives = 41/61 (67%), Gaps = 3/61 (4%)
Query 3 SPET---LHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNL 59
+PE +HQ ILFSDRGTP YR MHGYS H +K N G HY + H+KT+Q IKNL
Sbjct 183 TPENQVAIHQVMILFSDRGTPANYRSMHGYSGHTYKWSNKNGDWHYVQVHIKTDQGIKNL 242
Query 60 S 60
+
Sbjct 243 T 243
> YGR088w
Length=573
Score = 65.9 bits (159), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 44/71 (61%), Gaps = 0/71 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 60
+L+PE++HQ T +F DRGTP + M+ YS H+F +VN EG Y +FH+ ++ + L+
Sbjct 190 TLNPESIHQITYMFGDRGTPASWASMNAYSGHSFIMVNKEGKDTYVQFHVLSDTGFETLT 249
Query 61 AAFVFYLSSSH 71
LS SH
Sbjct 250 GDKAAELSGSH 260
> SPCC757.07c
Length=512
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 31/61 (50%), Positives = 40/61 (65%), Gaps = 1/61 (1%)
Query 1 SLSPETLHQTTILFSDRG-TPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNL 59
S + E++HQ ILFSD G TP YR M G+SSH +K VN +G +Y K+H TNQ K L
Sbjct 172 SQNAESIHQVMILFSDLGGTPYSYRFMDGFSSHTYKFVNDKGEFYYCKWHFITNQGTKGL 231
Query 60 S 60
+
Sbjct 232 T 232
> At1g20630
Length=492
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/60 (46%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 60
S PE+LH + LF D G P YRHM G + + L+N G HY KFH K IK LS
Sbjct 177 SHHPESLHMFSFLFDDLGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLS 236
> At4g35090
Length=492
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 1 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNL 59
S PE+L+ T LF D G P YRHM G + + L+N G HY KFH K +K+L
Sbjct 177 SHHPESLNMFTFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSL 235
> At1g20620
Length=531
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 4 PETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS--- 60
PE+L +F D G P YRHM G+ H + L+ G V + KFH K IKNL+
Sbjct 219 PESLLTWCWMFDDVGIPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLTDEE 278
Query 61 AAFVFYLSSSHA 72
A V + SHA
Sbjct 279 AKVVGGANHSHA 290
> CE25017
Length=611
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 38 NSEGVVHYAKFHLKTNQKIKNLSAAFVFYLSSSHAVSFRRSTAPARRR 85
++EGVV + + N+KIKN FV ++ A +R A + R
Sbjct 304 HAEGVVDVIVYSVPDNEKIKNRGFCFVDFIDHKTASDIKRKIAQHKIR 351
> Hs4504335
Length=1663
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 26/57 (45%), Gaps = 0/57 (0%)
Query 29 YSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSAAFVFYLSSSHAVSFRRSTAPARRR 85
Y + A + + +H+ + LK ++K ++A+ FY +S S S RR
Sbjct 972 YDADAIRECAEKVALHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYRR 1028
> Hs22047434
Length=381
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 26/64 (40%), Gaps = 10/64 (15%)
Query 38 NSEGVVHYAKFHLKTNQKIKNLSAAFVFYLSSSHAVSFRRSTAPARRRRASQASSLCAAA 97
SEG A +HLKT LS +HA+ +AP A A + A A
Sbjct 241 KSEGSRLEAVWHLKT----------LELLLSGTHALPIHLPSAPGAPADAEDAGNDSAPA 290
Query 98 LAIG 101
L +G
Sbjct 291 LVVG 294
Lambda K H
0.322 0.128 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1185472426
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40