bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6463_orf1
Length=142
Score E
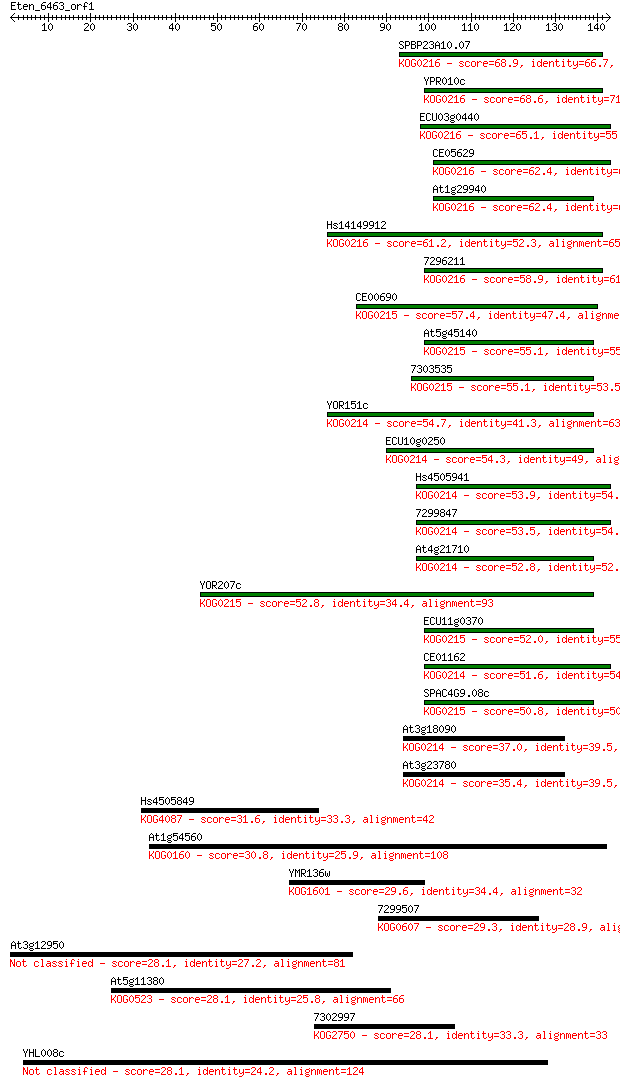
Sequences producing significant alignments: (Bits) Value
SPBP23A10.07 68.9 3e-12
YPR010c 68.6 3e-12
ECU03g0440 65.1 5e-11
CE05629 62.4 3e-10
At1g29940 62.4 3e-10
Hs14149912 61.2 5e-10
7296211 58.9 3e-09
CE00690 57.4 8e-09
At5g45140 55.1 4e-08
7303535 55.1 4e-08
YOR151c 54.7 5e-08
ECU10g0250 54.3 7e-08
Hs4505941 53.9 1e-07
7299847 53.5 1e-07
At4g21710 52.8 2e-07
YOR207c 52.8 2e-07
ECU11g0370 52.0 4e-07
CE01162 51.6 5e-07
SPAC4G9.08c 50.8 7e-07
At3g18090 37.0 0.012
At3g23780 35.4 0.036
Hs4505849 31.6 0.47
At1g54560 30.8 0.90
YMR136w 29.6 1.8
7299507 29.3 2.3
At3g12950 28.1 5.1
At5g11380 28.1 5.4
7302997 28.1 5.6
YHL008c 28.1 6.0
> SPBP23A10.07
Length=1227
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/48 (66%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 93 VPVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTP 140
VP +VE SPT LS+ A +TPFS+ NQSPRN+YQCQM KQTMGTP
Sbjct 720 VPKVSTHVEYSPTNVLSIVANMTPFSDFNQSPRNMYQCQMGKQTMGTP 767
> YPR010c
Length=1203
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 30/42 (71%), Positives = 35/42 (83%), Gaps = 0/42 (0%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTP 140
+VE +PT LS+ A LTPFS+ NQSPRN+YQCQM KQTMGTP
Sbjct 688 HVEFTPTNILSILANLTPFSDFNQSPRNMYQCQMGKQTMGTP 729
> ECU03g0440
Length=1062
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 25/45 (55%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 98 DYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTPFH 142
DY E+ + L + A L P+S+HNQSPRN+YQCQM KQ +G P H
Sbjct 619 DYYEIDKSGILGIIAGLIPYSDHNQSPRNMYQCQMAKQAIGHPAH 663
> CE05629
Length=1127
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/42 (61%), Positives = 29/42 (69%), Gaps = 0/42 (0%)
Query 101 ELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTPFH 142
EL P+ S L PF +HNQSPRN+YQCQM KQTMGT H
Sbjct 639 ELHPSCLFSFAGNLIPFPDHNQSPRNVYQCQMGKQTMGTAVH 680
> At1g29940
Length=1114
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/38 (68%), Positives = 31/38 (81%), Gaps = 0/38 (0%)
Query 101 ELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
E+ PT +SV A LTP+S+HNQSPRN+YQCQM KQTM
Sbjct 614 EIHPTGMISVVANLTPWSDHNQSPRNMYQCQMAKQTMA 651
> Hs14149912
Length=459
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/65 (52%), Positives = 41/65 (63%), Gaps = 7/65 (10%)
Query 76 MDVRGFQEVRDALVTSHVPVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQ 135
M+V F++ A VT+H EL P + LSV A PFS+ NQSPRN+YQCQM KQ
Sbjct 1 MNVAIFEDEVFAGVTTHQ-------ELFPHSLLSVIANFIPFSDLNQSPRNMYQCQMGKQ 53
Query 136 TMGTP 140
TMG P
Sbjct 54 TMGFP 58
> 7296211
Length=1129
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 26/42 (61%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTP 140
++EL+ T F+S A L P ++NQSPRN+YQCQM KQTMGTP
Sbjct 656 HLELAKTHFMSNLANLIPMPDYNQSPRNMYQCQMGKQTMGTP 697
> CE00690
Length=1207
Score = 57.4 bits (137), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 36/60 (60%), Gaps = 3/60 (5%)
Query 83 EVRDALVTSH---VPVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGT 139
E+ DAL+ + + ++E+ P L V A L P+ +HNQSPRN YQC M KQ MGT
Sbjct 703 EMNDALIAVYGREIGPETTHLEIEPFTLLGVCAGLIPYPHHNQSPRNTYQCAMGKQAMGT 762
> At5g45140
Length=1194
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 22/40 (55%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
++E+ P L V A L P+ +HNQSPRN YQC M KQ MG
Sbjct 688 HIEIEPFTILGVVAGLIPYPHHNQSPRNTYQCAMGKQAMG 727
> 7303535
Length=1137
Score = 55.1 bits (131), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 96 RYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
R ++E+ P L V A L P+ +HNQSPRN YQC M KQ MG
Sbjct 660 RTTHLEIEPFTLLGVCAGLVPYPHHNQSPRNTYQCAMGKQAMG 702
> YOR151c
Length=1224
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Query 76 MDVRGFQEVRDALVTSHVPVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQ 135
+DV + +R SH + + E+ P+ L V A++ PF +HNQSPRN YQ M KQ
Sbjct 721 LDVDPAKRIR----VSHHATTFTHCEIHPSMILGVAASIIPFPDHNQSPRNTYQSAMGKQ 776
Query 136 TMG 138
MG
Sbjct 777 AMG 779
> ECU10g0250
Length=1141
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 90 TSHVPVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
+S V V Y + E+ P L + A+ PF +HNQSPRN YQ M KQ MG
Sbjct 657 SSDVSVSYTHCEIHPALILGICASTIPFPDHNQSPRNTYQSAMGKQAMG 705
> Hs4505941
Length=1174
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 31/49 (63%), Gaps = 3/49 (6%)
Query 97 YDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG---TPFH 142
Y + E+ P+ L V A++ PF +HNQSPRN YQ M KQ MG T FH
Sbjct 693 YTHCEIHPSMILGVCASIIPFPDHNQSPRNTYQSAMGKQAMGVYITNFH 741
> 7299847
Length=1176
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 97 YDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG---TPFH 142
Y + E+ P L V A++ PF +HNQSPRN YQ M KQ MG T FH
Sbjct 695 YTHCEIHPAMILGVCASIIPFPDHNQSPRNTYQSAMGKQAMGVYITNFH 743
> At4g21710
Length=1188
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 97 YDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
Y + E+ P+ L V A++ PF +HNQSPRN YQ M KQ MG
Sbjct 701 YTHCEIHPSLILGVCASIIPFPDHNQSPRNTYQSAMGKQAMG 742
> YOR207c
Length=1149
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 32/98 (32%), Positives = 48/98 (48%), Gaps = 5/98 (5%)
Query 46 VVQDNQGRLESDY-QSLFNGQCD-SSTPNCGRMDVRGFQEVRD---ALVTSHVPVRYDYV 100
+V D Q R++ + + L +G+ D G ++ E D AL + ++
Sbjct 614 IVSDGQSRVKDIHLRKLLDGELDFDDFLKLGLVEYLDVNEENDSYIALYEKDIVPSMTHL 673
Query 101 ELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
E+ P L A L P+ +HNQSPRN YQC M KQ +G
Sbjct 674 EIEPFTILGAVAGLIPYPHHNQSPRNTYQCAMGKQAIG 711
> ECU11g0370
Length=1110
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/40 (55%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
++E+S A L A L PF +HNQSPRN YQC M KQ +G
Sbjct 663 HLEISEFAILGYVAGLVPFPHHNQSPRNTYQCAMGKQAIG 702
> CE01162
Length=1193
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 29/47 (61%), Gaps = 3/47 (6%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG---TPFH 142
+ E+ P L V A++ PF +HNQSPRN YQ M KQ MG T FH
Sbjct 701 HCEIHPAMILGVCASIIPFPDHNQSPRNTYQSAMGKQAMGVYTTNFH 747
> SPAC4G9.08c
Length=1165
Score = 50.8 bits (120), Expect = 7e-07, Method: Composition-based stats.
Identities = 20/40 (50%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 99 YVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMG 138
++E+ P L A L P+ +HNQSPRN YQC M KQ +G
Sbjct 685 HLEIEPFTILGAVAGLIPYPHHNQSPRNTYQCAMGKQAIG 724
> At3g18090
Length=1038
Score = 37.0 bits (84), Expect = 0.012, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 94 PVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQ 131
P Y + EL + L V+ A+ PF+NH+ R +YQ Q
Sbjct 548 PKNYTHCELDLSFLLGVSCAIVPFANHDHGKRVLYQSQ 585
> At3g23780
Length=946
Score = 35.4 bits (80), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 94 PVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQ 131
P Y + EL + L V+ A+ PF+NH+ R +YQ Q
Sbjct 457 PKIYTHCELDLSFLLGVSCAVVPFANHDHGRRVLYQSQ 494
> Hs4505849
Length=144
Score = 31.6 bits (70), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 16/42 (38%), Gaps = 0/42 (0%)
Query 32 LRDQIAEVDNTEDTVVQDNQGRLESDYQSLFNGQCDSSTPNC 73
R Q+ E D T N+ YQ N C STP C
Sbjct 103 CRSQLCECDKAAATCFARNKTTYNKKYQYYSNKHCRGSTPRC 144
> At1g54560
Length=1529
Score = 30.8 bits (68), Expect = 0.90, Method: Composition-based stats.
Identities = 28/108 (25%), Positives = 47/108 (43%), Gaps = 6/108 (5%)
Query 34 DQIAEVDNTEDTVVQDNQGRLESDYQSLFNGQCDSSTPNCGRMDVRGFQEVRDALVTSHV 93
D + E N +D VQ G+ + S + D P G D+ + + V ++
Sbjct 27 DGLVEKINGQDVEVQATNGKKITAKLSKIYPK-DMEAPAGGVDDMTKLSYLHEPGVLQNL 85
Query 94 PVRYDYVELSPTAFLSVTAALTPFSNHNQSPRNIYQCQMLKQTMGTPF 141
+RY+ E+ T ++ A+ PF Q +IY M++Q G PF
Sbjct 86 KIRYELNEIY-TYTGNILIAINPF----QRLPHIYDAHMMQQYKGAPF 128
> YMR136w
Length=560
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 67 DSSTPNCGRMDVRGFQEVRDALVTSHVPVRYD 98
+SS+P ++ + QE+ D++ T H+P YD
Sbjct 289 ESSSPFSNKIKLPSLQELTDSISTQHLPTFYD 320
> 7299507
Length=1142
Score = 29.3 bits (64), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 88 LVTSHVPVRYDYVELSPTAFLSVTAALTPFSNHNQSPR 125
+V H +RYDY++ P + A + +S+ N +P+
Sbjct 451 VVLQHFSMRYDYMKERPNIYQPSQAYMDAYSDENYNPK 488
> At3g12950
Length=558
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 2/83 (2%)
Query 1 AMVMLVATRSTGHVLLNERSMTAHQPLIEKILRDQIAEVDNTEDTVVQDNQGRLESDYQS 60
M +VA S +V L + + + L + Q+ +D E + +E +
Sbjct 440 GMSSMVADSSPPYVNLKKEKRSPEEKLEASLGPLQVQHIDLEERIETKGGAPSVEHQFMP 499
Query 61 LFNGQCDSST-PNCGRMD-VRGF 81
F+GQC +S P R D V GF
Sbjct 500 TFSGQCSASAWPETAREDLVAGF 522
> At5g11380
Length=700
Score = 28.1 bits (61), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 25 QPLIEKILRDQIAEVDNTEDTVVQDNQGRLESDYQSLFNGQ-CDSSTPNCGRMDVRGFQE 83
Q K+L + + + + +++ + RLES+Y S G C+S + G R +
Sbjct 149 QTYAHKVLTRRWSAIPSRQNSGISGVTSRLESEYDSFGTGHGCNSISAGLGLAVARDMKG 208
Query 84 VRDALVT 90
RD +V
Sbjct 209 KRDRVVA 215
> 7302997
Length=987
Score = 28.1 bits (61), Expect = 5.6, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 73 CGRMDVRGFQEVRDALVTSHVPVRYDYVELSPT 105
CG++ V G++ R +T + P ++V +SPT
Sbjct 493 CGQISVNGYRARRQEAITIYSPKGLNWVSISPT 525
> YHL008c
Length=627
Score = 28.1 bits (61), Expect = 6.0, Method: Composition-based stats.
Identities = 30/128 (23%), Positives = 57/128 (44%), Gaps = 8/128 (6%)
Query 4 MLVATRSTGHVLLNERSMTAHQP---LIEKILRDQIAEVDNTEDTVVQDNQGRLESDYQS 60
++V R + L E QP + +++R Q E D+ +D + E + S
Sbjct 262 LVVVERDRKRLSLPEYEARDEQPELNMDSRVVRIQKNECDDDATETGEDLENLTEKGFAS 321
Query 61 LFNGQCDSSTPNCGRMDVRGFQEVRDALVTS-HVPVRYDYVELSPTAFLSVTAALTPFSN 119
++N D+S+ GR + + + +++TS +V + D E P F+ + + T S
Sbjct 322 IYNTNHDNSSYFTGR-SLNSLRSIPSSVITSDNVTMESDLGE--PVQFIPKSNSTTR-SP 377
Query 120 HNQSPRNI 127
H P N+
Sbjct 378 HLGLPHNL 385
Lambda K H
0.318 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40