bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6457_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
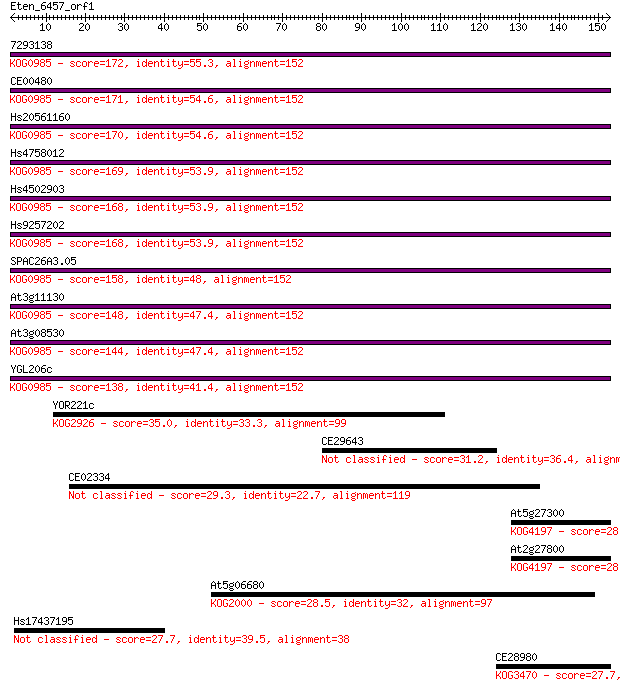
7293138 172 2e-43
CE00480 171 7e-43
Hs20561160 170 1e-42
Hs4758012 169 2e-42
Hs4502903 168 4e-42
Hs9257202 168 4e-42
SPAC26A3.05 158 3e-39
At3g11130 148 4e-36
At3g08530 144 9e-35
YGL206c 138 5e-33
YOR221c 35.0 0.051
CE29643 31.2 0.87
CE02334 29.3 2.7
At5g27300 28.9 4.0
At2g27800 28.9 4.3
At5g06680 28.5 5.3
Hs17437195 27.7 9.2
CE28980 27.7 9.2
> 7293138
Length=1678
Score = 172 bits (436), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 84/153 (54%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE+YT+L D+KR ++ T ++ EW+ FFG +S + +E L ML ++
Sbjct 618 CEKAGLLQRALEHYTDLYDIKRAVVHTH-MLNAEWLVSFFGTLSVEDSLECLKAMLTANL 676
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V +A K+HEQL + LI +FE F SY+G+FYFL SI+ FS DPEVHFKYI+AA
Sbjct 677 RQNLQICVQIATKYHEQLTNKALIDLFEGFKSYDGLFYFLSSIVNFSQDPEVHFKYIQAA 736
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K N +EVER+CRE Y P+RVK FLK+A+L
Sbjct 737 CKTNQIKEVERICRESNCYNPERVKNFLKEAKL 769
> CE00480
Length=1681
Score = 171 bits (432), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 111/153 (72%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T + +W+ +FG +S + VE L ML +
Sbjct 619 CEKAGLLQRALEHFTDLYDIKRTVVHTH-LLKPDWLVGYFGSLSVEDSVECLKAMLTQNI 677
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ VV +A K+HEQLG ++LI MFE SYEG+FYFLGSI+ FS DPEVHFKYI+AA
Sbjct 678 RQNLQVVVQIASKYHEQLGADKLIEMFENHKSYEGLFYFLGSIVNFSQDPEVHFKYIQAA 737
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
+ +EVER+CRE + Y+ +RVK FLK+A+L
Sbjct 738 TRTGQIKEVERICRESQCYDAERVKNFLKEAKL 770
> Hs20561160
Length=1285
Score = 170 bits (430), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL Q+ALE+YT+L D+KR ++ T ++ EW+ FFG +S + VE L ML ++
Sbjct 617 CEKAGLLQQALEHYTDLYDIKRAVVHTH-LLNPEWLVNFFGSLSVEDSVECLHAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQLG + L+ +FE F SY+G+FYFLGSI+ FS DP+VH KYI+AA
Sbjct 676 RQNLQLCVQVASKYHEQLGTQALVELFESFKSYKGLFYFLGSIVNFSQDPDVHLKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESSCYNPERVKNFLKEAKL 768
> Hs4758012
Length=1675
Score = 169 bits (429), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 82/153 (53%), Positives = 111/153 (72%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T ++ EW+ +FG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHFTDLYDIKRAVVHTH-LLNPEWLVNYFGSLSVEDSLECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL + LI +FE F S+EG+FYFLGSI+ FS DP+VHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTQSLIELFESFKSFEGLFYFLGSIVNFSQDPDVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y+P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESNCYDPERVKNFLKEAKL 768
> Hs4502903
Length=1569
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 82/153 (53%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL Q+ALE+YT+L D+KR ++ T ++ EW+ FFG +S + VE L ML ++
Sbjct 617 CEKAGLLQQALEHYTDLYDIKRAVVHTH-LLNPEWLVNFFGSLSVEDSVECLHAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+H+QLG + L+ +FE F SY+G+FYFLGSI+ FS DP+VH KYI+AA
Sbjct 676 RQNLQLCVQVASKYHKQLGTQALVELFESFKSYKGLFYFLGSIVNFSQDPDVHLKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESSCYNPERVKNFLKEAKL 768
> Hs9257202
Length=1626
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 82/153 (53%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL Q+ALE+YT+L D+KR ++ T ++ EW+ FFG +S + VE L ML ++
Sbjct 617 CEKAGLLQQALEHYTDLYDIKRAVVHTH-LLNPEWLVNFFGSLSVEDSVECLHAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+H+QLG + L+ +FE F SY+G+FYFLGSI+ FS DP+VH KYI+AA
Sbjct 676 RQNLQLCVQVASKYHKQLGTQALVELFESFKSYKGLFYFLGSIVNFSQDPDVHLKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESSCYNPERVKNFLKEAKL 768
> SPAC26A3.05
Length=1666
Score = 158 bits (400), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 73/153 (47%), Positives = 109/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE Y + AD+KRV++ + ++ EW+ +F R SPD + L +MLRS+
Sbjct 614 CERAGLVQRALELYDKPADIKRVIVHSN-LLNPEWLMNYFSRFSPDEVYDYLREMLRSNL 672
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ VV +A ++ + +G +R+I MFE+F ++EG++Y+LGSI+ + DPEV +KYI+AA
Sbjct 673 RQNLQIVVQIATRYSDLVGAQRIIEMFEKFKTFEGLYYYLGSIVNITEDPEVVYKYIQAA 732
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
+N EVER+CR+ Y P++VK LK+A+L
Sbjct 733 CLMNQFTEVERICRDNNVYNPEKVKNLLKEAKL 765
> At3g11130
Length=1705
Score = 148 bits (373), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 72/153 (47%), Positives = 109/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE AGLY ++L++Y+EL D+KRV++ T I + + +FFG +S + +E + D+L +
Sbjct 631 CEKAGLYIQSLKHYSELPDIKRVIVNTHA-IEPQALVEFFGTLSSEWAMECMKDLLLVNL 689
Query 61 Q-NLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
+ NLQ +V V ++ EQLG++ I +FE+F SYEG+++FLGS L+ S DPE+HFKYIEAA
Sbjct 690 RGNLQIIVQVCAEYCEQLGVDACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYIEAA 749
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
AK +EVERV RE +Y+ ++ K FL +A+L
Sbjct 750 AKTGQIKEVERVTRESNFYDAEKTKNFLMEAKL 782
> At3g08530
Length=1516
Score = 144 bits (362), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 72/156 (46%), Positives = 109/156 (69%), Gaps = 5/156 (3%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE AGLY ++L++Y+EL D+KRV++ T I + + +FFG +S + +E + D+L +
Sbjct 441 CEKAGLYIQSLKHYSELPDIKRVIVNTHA-IEPQALVEFFGTLSSEWAMECMKDLLLVNL 499
Query 61 Q-NLQAVV---GVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYI 116
+ NLQ +V G ++ EQLG++ I +FE+F SYEG+++FLGS L+ S DPE+HFKYI
Sbjct 500 RGNLQIIVQASGACKEYCEQLGVDACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYI 559
Query 117 EAAAKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
EAAAK +EVERV RE +Y+ ++ K FL +A+L
Sbjct 560 EAAAKTGQIKEVERVTRESNFYDAEKTKNFLMEAKL 595
> YGL206c
Length=1653
Score = 138 bits (347), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 63/153 (41%), Positives = 104/153 (67%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
E AGLYQRALENYT++ D+KR ++ T + +W+ +FG+++ + + L ++ ++
Sbjct 623 SEKAGLYQRALENYTDIKDIKRCVVHTNA-LPIDWLVGYFGKLNVEQSLACLKALMDNNI 681
Query 61 Q-NLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
Q N+Q VV VA KF + +G LI +FE +++ EG++Y+L S++ + D +V +KYIEAA
Sbjct 682 QANIQTVVQVATKFSDLIGPSTLIKLFEDYNATEGLYYYLASLVNLTEDKDVVYKYIEAA 741
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
AK+ +E+ER+ ++ Y+P+RVK FLK A L
Sbjct 742 AKMKQYREIERIVKDNNVYDPERVKNFLKDANL 774
> YOR221c
Length=384
Score = 35.0 bits (79), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 48/105 (45%), Gaps = 17/105 (16%)
Query 12 ENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSSQNLQAVVGVAI 71
ENY L +K K+S +MA FFGR I + IL ++R+ S+ Q ++
Sbjct 9 ENYRPLNILKM------QKVSSSYMA-FFGRQGTSISISILKAIIRNKSREFQTILSQNG 61
Query 72 KFHEQLGMERLIAMFERFSSYEGI------FYFLGSILAFSTDPE 110
K L L +F+ SS I FY L IL+ +DP+
Sbjct 62 KESNDL----LQYIFQNPSSPGSIAVCSNLFYQLYQILSNPSDPQ 102
> CE29643
Length=603
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 80 ERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIE-AAAKLN 123
E LI + +SY G +L++STDP +YI+ AK+N
Sbjct 238 EELIDLLTEENSYPPAMILRGKMLSYSTDPNEATRYIDKVGAKIN 282
> CE02334
Length=442
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 27/138 (19%), Positives = 62/138 (44%), Gaps = 30/138 (21%)
Query 16 ELADVKRVLLQTGGKISQ-------EWMAQFFGRMSPDICVEILTDMLRSSSQNLQAVV- 67
EL + ++++ Q G +I + +W A GR+ + E+ SS+++ V
Sbjct 15 ELPEARKIMKQIGAQIPRIVESIQVDWSA--LGRV---VAEEV------KSSEDMPPVAA 63
Query 68 ----GVAIKFHEQLGMERLIAMFERFSSYEG-------IFYFLGSILAFSTDPEVHFKYI 116
G A+ H+ +G+++++ + + Y+G + G I+ D + +Y
Sbjct 64 STKDGYAVIAHDGIGLKKMVGVSLAGNIYQGAVEIGKCVRISTGGIIPEGADAVIMREYT 123
Query 117 EAAAKLNHTQEVERVCRE 134
E + ++E E +C++
Sbjct 124 ELVRQDQQSEETEIICKQ 141
> At5g27300
Length=510
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 128 VERVCREGKYYEPQRVKEFLKQAQL 152
V+ CR+GKY E R+ E L++ QL
Sbjct 320 VDESCRKGKYDEATRLLEMLREKQL 344
> At2g27800
Length=427
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 128 VERVCREGKYYEPQRVKEFLKQAQL 152
V+ CR+GKY E R+ E L++ QL
Sbjct 386 VDESCRKGKYDEATRLLEMLREKQL 410
> At5g06680
Length=838
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 46/106 (43%), Gaps = 22/106 (20%)
Query 52 LTDMLRSSSQNLQAVVGVAIKFHE-QLGMERLIAMFE---RFSS-----YEGIFYFLGSI 102
L D+L + + L A+VG ++ + Q E L +FE RF S YEGI
Sbjct 694 LDDLLAAHEKYLNAIVGKSLLGEQSQTIRESLFVLFELILRFRSHADRLYEGIH------ 747
Query 103 LAFSTDPEVHFKYIEAAAKLNHTQEVERVCREGKYYEPQRVKEFLK 148
E+ + E+ + N +QE EG+ QR EFL+
Sbjct 748 -------ELQIRSKESGREKNKSQEPGSWISEGRKGLTQRAGEFLQ 786
> Hs17437195
Length=307
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 3/38 (7%)
Query 2 EAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQF 39
E +GL Q+ + E A + R LLQ+GGKI +++ F
Sbjct 127 EPSGLIQQEI---MEKAAISRCLLQSGGKIFPQYVLMF 161
> CE28980
Length=111
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 124 HTQEVERVCREGKYYEPQRVKEFLKQAQL 152
T V+R+ +E YYE Q VKE K AQL
Sbjct 14 KTGTVQRLVKEVAYYEKQVVKEEQKAAQL 42
Lambda K H
0.323 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40