bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6375_orf1
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
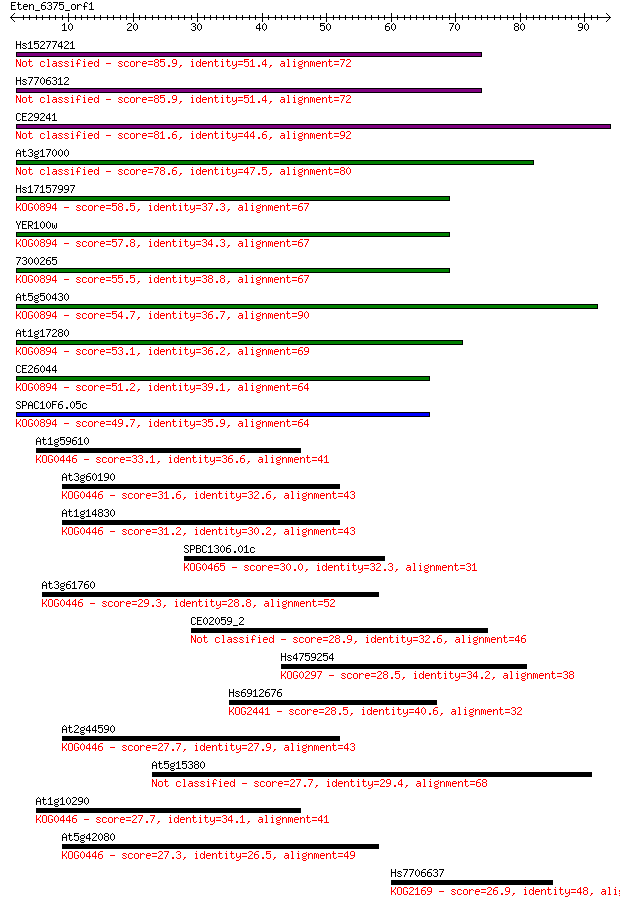
Hs15277421 85.9 2e-17
Hs7706312 85.9 2e-17
CE29241 81.6 3e-16
At3g17000 78.6 3e-15
Hs17157997 58.5 3e-09
YER100w 57.8 5e-09
7300265 55.5 3e-08
At5g50430 54.7 4e-08
At1g17280 53.1 1e-07
CE26044 51.2 4e-07
SPAC10F6.05c 49.7 1e-06
At1g59610 33.1 0.13
At3g60190 31.6 0.38
At1g14830 31.2 0.52
SPBC1306.01c 30.0 1.1
At3g61760 29.3 1.8
CE02059_2 28.9 2.1
Hs4759254 28.5 3.0
Hs6912676 28.5 3.5
At2g44590 27.7 4.8
At5g15380 27.7 4.9
At1g10290 27.7 5.0
At5g42080 27.3 7.0
Hs7706637 26.9 8.6
> Hs15277421
Length=318
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 0/72 (0%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF+V KKICLS S HHPE WQP+W IRT L A+ F PT +GA+ +L+ + ++RR +
Sbjct 81 NGRFEVGKKICLSISGHHPETWQPSWSIRTALLAIIGFMPTKGEGAIGSLDYTPEERRAL 140
Query 62 ALESASWVCPVC 73
A +S + C C
Sbjct 141 AKKSQDFCCEGC 152
> Hs7706312
Length=325
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 0/72 (0%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF+V KKICLS S HHPE WQP+W IRT L A+ F PT +GA+ +L+ + ++RR +
Sbjct 88 NGRFEVGKKICLSISGHHPETWQPSWSIRTALLAIIGFMPTKGEGAIGSLDYTPEERRAL 147
Query 62 ALESASWVCPVC 73
A +S + C C
Sbjct 148 AKKSQDFCCEGC 159
> CE29241
Length=314
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 53/92 (57%), Gaps = 10/92 (10%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF++NKK+CLS S +HPE W P+W IRT L AL F P+ GAL +L+ K+R+++
Sbjct 81 NGRFELNKKVCLSISGYHPETWLPSWSIRTALLALIGFLPSTPGGALGSLDYPPKERQRL 140
Query 62 ALESASWVCPVCKRPNRQLVEEGCAAPTDTLP 93
A S W C C GC T LP
Sbjct 141 AKLSCEWKCKEC----------GCVMKTALLP 162
> At3g17000
Length=309
Score = 78.6 bits (192), Expect = 3e-15, Method: Composition-based stats.
Identities = 38/80 (47%), Positives = 52/80 (65%), Gaps = 1/80 (1%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF+ N KICLS S++HPE WQP+W +RT L AL AF PT GAL +++ + +RR +
Sbjct 83 NGRFETNTKICLSISNYHPEHWQPSWSVRTALVALIAFMPTSPNGALGSVDYPKDERRTL 142
Query 62 ALESASWVCPVCKRPNRQLV 81
A++S P P RQ +
Sbjct 143 AIKSRE-TPPKYGSPERQKI 161
> Hs17157997
Length=224
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 41/67 (61%), Gaps = 1/67 (1%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF N ++CLS + HP+ W PAW + T+L L +F L ++E S+ +RQ+
Sbjct 49 NGRFKCNTRLCLSITDFHPDTWNPAWSVSTILTGLLSFMVEKGP-TLGSIETSDFTKRQL 107
Query 62 ALESASW 68
A++S ++
Sbjct 108 AVQSLAF 114
> YER100w
Length=250
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
NGRF N ++CLS S +HP+ W P W + T+L+ L +F T + ++ S+ ++ +
Sbjct 77 NGRFKPNTRLCLSMSDYHPDTWNPGWSVSTILNGLLSFM-TSDEATTGSITTSDHQKKTL 135
Query 62 ALESASW 68
A S S+
Sbjct 136 ARNSISY 142
> 7300265
Length=251
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFF--PTPAQGALHALEASEKDRR 59
NGRF N ++CLS S HP+ W P W + T+L L +F TP G ++E+S D++
Sbjct 86 NGRFKTNTRLCLSISDFHPDTWNPTWCVGTILTGLLSFMLESTPTLG---SIESSNYDKQ 142
Query 60 QMALESASW 68
A +S ++
Sbjct 143 MFAQKSLAF 151
> At5g50430
Length=243
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 50/103 (48%), Gaps = 16/103 (15%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFF--PTPAQGALHALEASEKDRR 59
NGRF KKICLS S HPE W P W + ++L L +F +P G+++ S +++
Sbjct 77 NGRFVTQKKICLSMSDFHPESWNPMWSVSSILTGLLSFMMDNSPTTGSVN---TSVAEKQ 133
Query 60 QMALESASWVCP-----------VCKRPNRQLVEEGCAAPTDT 91
++A S ++ C V K +Q+ EE A T
Sbjct 134 RLAKSSLAFNCKSVTFRKLFPEYVEKYSQQQVAEEEAATQQTT 176
> At1g17280
Length=237
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 5/71 (7%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFF--PTPAQGALHALEASEKDRR 59
NGRF KKICLS S HPE W P W + ++L L +F +P G+++ + +++
Sbjct 77 NGRFMTQKKICLSMSDFHPESWNPMWSVSSILTGLLSFMMDTSPTTGSVN---TTVIEKQ 133
Query 60 QMALESASWVC 70
++A S ++ C
Sbjct 134 RLAKSSLAFNC 144
> CE26044
Length=218
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 37/66 (56%), Gaps = 5/66 (7%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFF--PTPAQGALHALEASEKDRR 59
NGRF N ++CLS S +HPE W P W + +L L +F +PA G ++ + +D+R
Sbjct 91 NGRFQTNTRLCLSISDYHPESWNPGWTVSAILIGLHSFMNENSPAAG---SIAGTPQDQR 147
Query 60 QMALES 65
A S
Sbjct 148 MYAAAS 153
> SPAC10F6.05c
Length=227
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 2 NGRFDVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQM 61
+GRF N ++CLS S HP+ W P+W + T+L L +F T + + SE RR
Sbjct 77 SGRFQTNTRLCLSFSDFHPKSWNPSWMVSTILVGLVSFM-TSDEITTGGIVTSESTRRTY 135
Query 62 ALES 65
A ++
Sbjct 136 AKDT 139
> At1g59610
Length=920
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query 5 FDVN--KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQ 45
FD+N K+I L A + P L P G+R+L+ + PA+
Sbjct 384 FDLNNVKRIVLEADGYQPYLISPEKGLRSLIKTVLELAKDPAR 426
> At3g60190
Length=621
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHAL 51
KKI A + P L P G R L++ +F PA+ ++ A+
Sbjct 390 KKIVSEADGYQPHLIAPEQGYRRLIEGALGYFRGPAEASVDAV 432
> At1g14830
Length=614
Score = 31.2 bits (69), Expect = 0.52, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHAL 51
+K+ A + P L P G R L+D ++F PA+ + A+
Sbjct 388 QKVVSEADGYQPHLIAPEQGYRRLIDGSISYFKGPAEATVDAV 430
> SPBC1306.01c
Length=558
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 10/31 (32%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 28 GIRTLLDALCAFFPTPAQGALHALEASEKDR 58
G++++LDA+C + P P++ AL A++ ++
Sbjct 125 GVQSVLDAVCDYLPNPSEVENIALNAADSEK 155
> At3g61760
Length=627
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 6 DVNKKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKD 57
D +K+ A + P L P G R L+++ PA+ A+ A+ + KD
Sbjct 401 DNVRKLITEADGYQPHLIAPEQGYRRLIESCLVSIRGPAEAAVDAVHSILKD 452
> CE02059_2
Length=279
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 29 IRTLLDALCAFFPTPAQGALHALEASEKDRRQMALESASWVCPVCK 74
I TL +A+ FP+ Q L A+E R++ +E++S+ P+ K
Sbjct 40 IDTLSNAMFTRFPSILQQFLIAIEPPTAVNRELGIENSSYEDPLTK 85
> Hs4759254
Length=522
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 43 PAQGALHALEASEKDRRQMALESASWVCPVCKRPNRQL 80
P +G LH +E + Q E A CP C+RP ++
Sbjct 135 PNEGCLHKMELRHLEDHQAHCEFALMDCPQCQRPFQKF 172
> Hs6912676
Length=536
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 35 ALCAFFPTPAQGALHALEASEKDRRQMALESA 66
AL +F P P Q + LEA EK R Q + +++
Sbjct 2 ALTSFLPAPTQLSQDQLEAEEKARSQRSRQTS 33
> At2g44590
Length=613
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHAL 51
K+I + + P L P G R L++ F PA+ +++A+
Sbjct 388 KRIVSESDGYQPHLIAPELGYRRLIEGSLNHFRGPAEASVNAI 430
> At5g15380
Length=375
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 30/68 (44%), Gaps = 1/68 (1%)
Query 23 WQPAWGIRTLLDALCAFFPTPAQGALHALEASEKDRRQMALESASWVCPVCKRPNRQLVE 82
W P+W RT L+ L + ++ EA E+ + L+ WV CK+ N V
Sbjct 130 WWPSWDGRTKLNCLLTCIAS-SRLTEKIREALERYDGETPLDVQKWVMYECKKWNLVWVG 188
Query 83 EGCAAPTD 90
+ AP D
Sbjct 189 KNKLAPLD 196
> At1g10290
Length=914
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query 5 FDVN--KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQ 45
FD+N K++ L A + P L P G+R+L+ + PA+
Sbjct 384 FDLNNVKRVVLEADGYQPYLISPEKGLRSLIKIVLELAKDPAR 426
> At5g42080
Length=610
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 9 KKICLSASSHHPELWQPAWGIRTLLDALCAFFPTPAQGALHALEASEKD 57
+K+ A + P L P G R L+++ PA+ ++ + A KD
Sbjct 387 RKLVTEADGYQPHLIAPEQGYRRLIESSIVSIRGPAEASVDTVHAILKD 435
> Hs7706637
Length=651
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 16/27 (59%), Gaps = 2/27 (7%)
Query 60 QMALESASWVCPVC--KRPNRQLVEEG 84
QM + +WVCPVC K P L+ +G
Sbjct 364 QMNEKKPTWVCPVCDKKAPYEHLIIDG 390
Lambda K H
0.321 0.133 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167934574
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40