bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6373_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
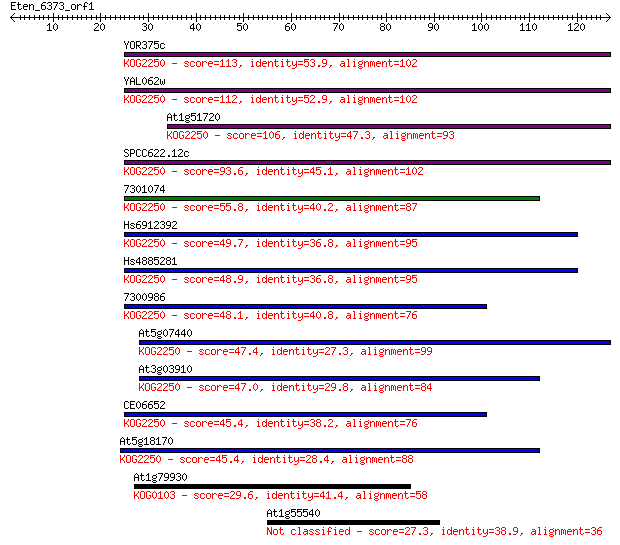
YOR375c 113 7e-26
YAL062w 112 2e-25
At1g51720 106 1e-23
SPCC622.12c 93.6 7e-20
7301074 55.8 2e-08
Hs6912392 49.7 1e-06
Hs4885281 48.9 2e-06
7300986 48.1 4e-06
At5g07440 47.4 7e-06
At3g03910 47.0 9e-06
CE06652 45.4 2e-05
At5g18170 45.4 2e-05
At1g79930 29.6 1.5
At1g55540 27.3 7.8
> YOR375c
Length=454
Score = 113 bits (283), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 55/104 (52%), Positives = 73/104 (70%), Gaps = 2/104 (1%)
Query 25 TTHKLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKD 84
+ +++R I +FM EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG LTGK
Sbjct 121 SNNEIRRICYAFMRELSRHIGQDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGVLTGKG 180
Query 85 VGWGGSEIRPEATGYGCAYFAENVLTKLLN--ESLRGKSCTLSG 126
+ WGGS IRPEATGYG Y+ + ++ N ES GK T+SG
Sbjct 181 LNWGGSLIRPEATGYGLVYYTQAMIDYATNGKESFEGKRVTISG 224
> YAL062w
Length=457
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/104 (51%), Positives = 73/104 (70%), Gaps = 2/104 (1%)
Query 25 TTHKLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKD 84
+ +++R I +FM EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG LTGK
Sbjct 122 SDNEIRRICYAFMRELSRHIGKDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGVLTGKG 181
Query 85 VGWGGSEIRPEATGYGCAYFAENVLTKLLN--ESLRGKSCTLSG 126
+ WGGS IRPEATG+G Y+ + ++ N ES GK T+SG
Sbjct 182 LNWGGSLIRPEATGFGLVYYTQAMIDYATNGKESFEGKRVTISG 225
> At1g51720
Length=624
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 44/93 (47%), Positives = 68/93 (73%), Gaps = 1/93 (1%)
Query 34 QSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKDVGWGGSEIR 93
QSFM E+ +++GP +D+P+ ++GVGTRE+G+LFGQY+RL+ F+G+ TG + W S +R
Sbjct 320 QSFMNEMYRYMGPDKDLPSEEVGVGTREMGYLFGQYRRLAGQFQGSFTGPRIYWAASSLR 379
Query 94 PEATGYGCAYFAENVLTKLLNESLRGKSCTLSG 126
EA+GYG YFA +L +N+ ++G C +SG
Sbjct 380 TEASGYGVVYFARLILAD-MNKEIKGLRCVVSG 411
> SPCC622.12c
Length=451
Score = 93.6 bits (231), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 69/104 (66%), Gaps = 2/104 (1%)
Query 25 TTHKLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKD 84
+ +++R Q+FM +L ++IGP DVPAGDIGV + +FG+YKRL + + G +TGK
Sbjct 124 SDNEIRRFSQAFMRQLFRYIGPQTDVPAGDIGVTGFVVMHMFGEYKRLRNEYSGVVTGKH 183
Query 85 VGWGGSEIRPEATGYGCAYFAENVLTKLLN--ESLRGKSCTLSG 126
+ GGS IRPEATGYG Y+ ++++ E+L+GK +SG
Sbjct 184 MLTGGSNIRPEATGYGVVYYVKHMIEHRTKGAETLKGKRVAISG 227
> 7301074
Length=590
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 48/93 (51%), Gaps = 6/93 (6%)
Query 25 TTHKLRWIFQSFMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKR----LSSNFEG 78
+ H+L I + F ELAK IGP DVPA D+G G RE+ ++ Y + L N
Sbjct 187 SEHELEKITRRFTLELAKKGFIGPGVDVPAPDMGTGEREMSWIADTYAKTIGHLDINAHA 246
Query 79 ALTGKDVGWGGSEIRPEATGYGCAYFAENVLTK 111
+TGK + GG R ATG G + EN + +
Sbjct 247 CVTGKPINQGGIHGRVSATGRGVFHGLENFINE 279
> Hs6912392
Length=558
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 48/101 (47%), Gaps = 6/101 (5%)
Query 25 TTHKLRWIFQSFMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYK----RLSSNFEG 78
T ++L I + F ELAK IGP DVPA D+ G RE+ ++ Y N
Sbjct 194 TENELEKITRRFTMELAKKGFIGPGVDVPAPDMNTGEREMSWIADTYASTIGHYDINAHA 253
Query 79 ALTGKDVGWGGSEIRPEATGYGCAYFAENVLTKLLNESLRG 119
+TGK + GG R ATG G + EN + + S+ G
Sbjct 254 CVTGKPISQGGIHGRISATGRGVFHGIENFINQASYMSILG 294
> Hs4885281
Length=558
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 48/101 (47%), Gaps = 6/101 (5%)
Query 25 TTHKLRWIFQSFMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYK----RLSSNFEG 78
T ++L I + F ELAK IGP DVPA D+ G RE+ ++ Y N
Sbjct 194 TDNELEKITRRFTMELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYASTIGHYDINAHA 253
Query 79 ALTGKDVGWGGSEIRPEATGYGCAYFAENVLTKLLNESLRG 119
+TGK + GG R ATG G + EN + + S+ G
Sbjct 254 CVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSILG 294
> 7300986
Length=520
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 25 TTHKLRWIFQSFMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRL----SSNFEG 78
T +L+ I + + EL K IGP DVPA D+ G RE+ ++ QY++ N
Sbjct 158 TVDELQTITRRYTMELLKRNMIGPGIDVPAPDVNTGPREMSWIVDQYQKTFGYKDINSSA 217
Query 79 ALTGKDVGWGGSEIRPEATGYG 100
+TGK V GG R ATG G
Sbjct 218 IVTGKPVHNGGINGRHSATGRG 239
> At5g07440
Length=411
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 50/99 (50%), Gaps = 1/99 (1%)
Query 28 KLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKDVGW 87
+L + + F ++ IG DVPA D+G + + ++ +Y + + +TGK +
Sbjct 116 ELERLTRVFTQKIHDLIGIHTDVPAPDMGTNAQTMAWILDEYSKFHGHSPAVVTGKPIDL 175
Query 88 GGSEIRPEATGYGCAYFAENVLTKLLNESLRGKSCTLSG 126
GGS R ATG G + E +L + +S++G + + G
Sbjct 176 GGSLGREAATGRGVVFATEALLAE-YGKSIQGLTFVIQG 213
> At3g03910
Length=411
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 43/84 (51%), Gaps = 0/84 (0%)
Query 28 KLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGKDVGW 87
+L + + F ++ IG DVPA D+G G + + ++ +Y + + +TGK +
Sbjct 116 ELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHGHSPAVVTGKPIDL 175
Query 88 GGSEIRPEATGYGCAYFAENVLTK 111
GGS R ATG G + E +L +
Sbjct 176 GGSLGRDAATGRGVLFATEALLNE 199
> CE06652
Length=536
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 25 TTHKLRWIFQSFMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKR----LSSNFEG 78
T +++ I + E AK +GP DVPA D+G G RE+G++ Y + L +
Sbjct 172 TDYEIEKITRRIAIEFAKKGFLGPGVDVPAPDMGTGEREMGWIADTYAQTIGHLDRDASA 231
Query 79 ALTGKDVGWGGSEIRPEATGYG 100
+TGK + GG R ATG G
Sbjct 232 CITGKPIVSGGIHGRVSATGRG 253
> At5g18170
Length=411
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 44/88 (50%), Gaps = 0/88 (0%)
Query 24 VTTHKLRWIFQSFMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLSSNFEGALTGK 83
++ +L + + F ++ IG DVPA D+G G + + ++ +Y + +TGK
Sbjct 112 LSISELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHGYSPAVVTGK 171
Query 84 DVGWGGSEIRPEATGYGCAYFAENVLTK 111
+ GGS R ATG G + E +L +
Sbjct 172 PIDLGGSLGRDAATGRGVMFGTEALLNE 199
> At1g79930
Length=831
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 34/69 (49%), Gaps = 12/69 (17%)
Query 27 HKLRWIFQSFMTELAKHIGPSRDVPAGD--------IGVGTREI---GFLFGQYKRLSSN 75
H LR I ++ T LA I + D+P D IG + ++ GF GQ K LS
Sbjct 166 HPLRLIHETTATALAYGIYKT-DLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHA 224
Query 76 FEGALTGKD 84
F+ +L G+D
Sbjct 225 FDRSLGGRD 233
> At1g55540
Length=1804
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 55 IGVGTREIGFL---FGQYKRLSSNFEGALTGKDVGWGGS 90
IG G + +G + FGQ +++ + GA G G+GGS
Sbjct 1628 IGGGQQALGSVLGSFGQSRQIGAGLPGATFGSPTGFGGS 1666
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40