bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6305_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
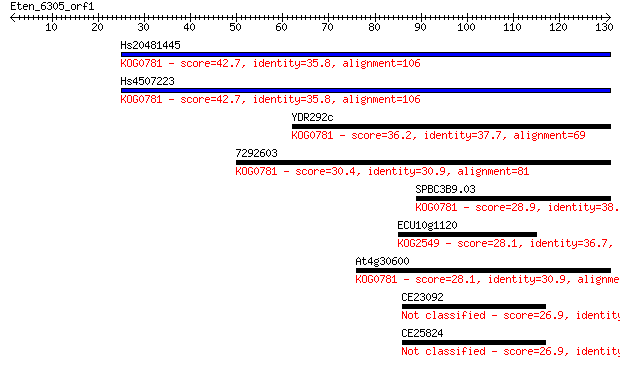
Hs20481445 42.7 2e-04
Hs4507223 42.7 2e-04
YDR292c 36.2 0.015
7292603 30.4 0.98
SPBC3B9.03 28.9 2.4
ECU10g1120 28.1 4.1
At4g30600 28.1 4.9
CE23092 26.9 8.5
CE25824 26.9 9.6
> Hs20481445
Length=638
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 51/115 (44%), Gaps = 10/115 (8%)
Query 25 EALDYSKKTGNG------DEAAHAIERKTYGA---DSDSQSSEEETEAAAAAAAATAEGS 75
E LDYS T NG E + I G D D SS++E A + + +G+
Sbjct 257 EVLDYSTPTTNGTPEAALSEDINLIRGTGSGGQLQDLDCSSSDDEGAAQNSTKPSATKGT 316
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
L + L+ G+K L D+E VL R H AKNVA ++ L SV L
Sbjct 317 -LGGMFGMLKGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKL 370
> Hs4507223
Length=638
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 50/115 (43%), Gaps = 10/115 (8%)
Query 25 EALDYSKKTGNG------DEAAHAIERKTYGA---DSDSQSSEEETEAAAAAAAATAEGS 75
E LDYS T NG E + I G D D SS++E A + +G+
Sbjct 257 EVLDYSTPTTNGTPEAALSEDINLIRGTGSGGQLQDLDCSSSDDEGAAQTLTKPSATKGT 316
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
L + L+ G+K L D+E VL R H AKNVA ++ L SV L
Sbjct 317 -LGGMFGMLKGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKL 370
> YDR292c
Length=621
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query 62 EAAAAAAAATAEGSWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDL 121
EA + +TA G FL K + GNK ++ESD++ VL Q KNVA E D
Sbjct 282 EAKNSGYVSTAFG----FLQKHV---LGNKTINESDLKSVLEKLTQQLITKNVAPEAADY 334
Query 122 LLLSVKKNL 130
L V +L
Sbjct 335 LTQQVSHDL 343
> 7292603
Length=631
Score = 30.4 bits (67), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 37/81 (45%), Gaps = 5/81 (6%)
Query 50 ADSDSQSSEEETEAAAAAAAATAEGSWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHF 109
AD++ SSE E E + G LS+ + G K + +D++ L R H
Sbjct 272 ADNEDASSEGEAEEQVQSKKGK-RGGLLSYF----KGIVGAKTMSLADLQPALEKMRDHL 326
Query 110 SAKNVAEEVTDLLLLSVKKNL 130
+KNVA E+ L SV +L
Sbjct 327 ISKNVASEIAAKLCDSVAASL 347
> SPBC3B9.03
Length=547
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 89 GNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
G K L E D+ +L ++H + KNVA + L SVK +L
Sbjct 252 GGKYLKEEDLSPILKQMQEHLTKKNVANSIALELCESVKASL 293
> ECU10g1120
Length=356
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 85 QKTAGNKVLDESDIEEVLPYFRQHFSAKNV 114
++TA + +ES I++++PYF HF+ + V
Sbjct 184 EQTAMECLHNESGIQQLVPYFVHHFNEQIV 213
> At4g30600
Length=634
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
W S + Q G L+ +D+ L ++ KNVAEE+ + L SV+ +L
Sbjct 329 WFSSV---FQSITGKANLERTDLGPALKALKERLMTKNVAEEIAEKLCESVEASL 380
> CE23092
Length=2129
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 86 KTAGNKVLDESDIEEVLPYFRQHFSAKNVAE 116
KT ++D++ IE + P + +HF AKN +
Sbjct 1431 KTRMKTLIDKATIERLWPGYEEHFYAKNTGK 1461
> CE25824
Length=754
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 86 KTAGNKVLDESDIEEVLPYFRQHFSAKNVAE 116
KT ++D++ IE + P + +HF AKN +
Sbjct 56 KTRMKTLIDKATIERLWPGYEEHFYAKNTGK 86
Lambda K H
0.303 0.119 0.321
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40