bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6283_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
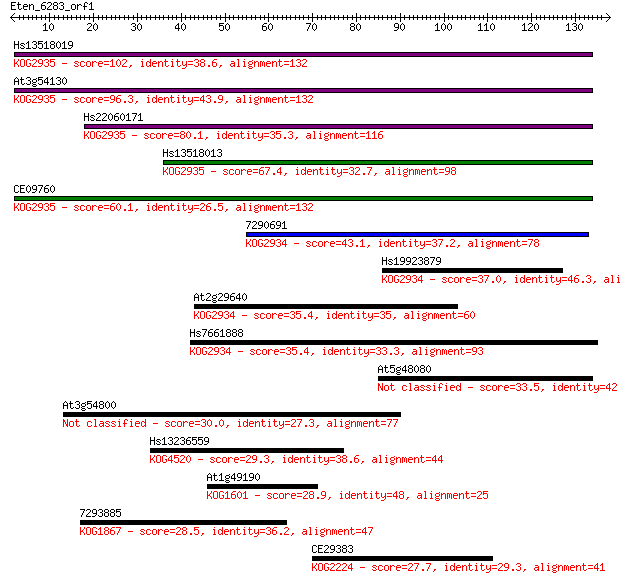
Hs13518019 102 2e-22
At3g54130 96.3 1e-20
Hs22060171 80.1 1e-15
Hs13518013 67.4 7e-12
CE09760 60.1 1e-09
7290691 43.1 2e-04
Hs19923879 37.0 0.010
At2g29640 35.4 0.028
Hs7661888 35.4 0.030
At5g48080 33.5 0.12
At3g54800 30.0 1.3
Hs13236559 29.3 2.6
At1g49190 28.9 3.4
7293885 28.5 3.7
CE29383 27.7 7.2
> Hs13518019
Length=361
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/136 (37%), Positives = 84/136 (61%), Gaps = 5/136 (3%)
Query 2 ELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHC 61
ELSSI +LD++ER MAEGG S+DY+ FL + SGN+ G+F++ V+ L+ +
Sbjct 32 ELSSIAHQLDEEERMRMAEGGVTSEDYRTFLQQPSGNMDDSGFFSIQVISNALKVWGLEL 91
Query 62 ICQRNVGEHEEAGVD----FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEA 117
I N E++ +D +I N+ +HW +R++ W+NL+S+ P +IS +L
Sbjct 92 IL-FNSPEYQRLRIDPINERSFICNYKEHWFTVRKLGKQWFNLNSLLTGPELISDTYLAL 150
Query 118 FLSSLKGQGYTIFIVR 133
FL+ L+ +GY+IF+V+
Sbjct 151 FLAQLQQEGYSIFVVK 166
> At3g54130
Length=280
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 58/140 (41%), Positives = 82/140 (58%), Gaps = 12/140 (8%)
Query 2 ELSSIGLELDQKERQLMAEG----GTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRK 57
+L+++ +LD KERQ+M EG G D FLAE S NV+ G F++ VL + L
Sbjct 38 DLAAVAADLDGKERQVMLEGAAVGGFAPGD---FLAEESHNVSLGGDFSIQVLQKALEVW 94
Query 58 RIHCICQRNVGEHEEAGVD----FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSF 113
+ I N + E A +D +I + DHW +R+V+G WYN DS+ AP +S F
Sbjct 95 DLQVI-PLNCPDAEPAQIDPELESAFICHLHDHWFCIRKVNGEWYNFDSLLAAPQHLSKF 153
Query 114 HLEAFLSSLKGQGYTIFIVR 133
+L AFL SLKG G++IFIV+
Sbjct 154 YLSAFLDSLKGAGWSIFIVK 173
> Hs22060171
Length=310
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 69/120 (57%), Gaps = 5/120 (4%)
Query 18 MAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVDF 77
MAEGG S++Y AFL + S N+ G+F++ V+ L+ + I N E+++ G+D
Sbjct 3 MAEGGVTSEEYLAFLQQPSENMDDTGFFSIQVISNALKFWGLEII-HFNNPEYQKLGIDP 61
Query 78 ----GYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEAFLSSLKGQGYTIFIVR 133
+I N+ HW +R+ W+NL+S+ P +IS L FL+ L+ Q Y++F+V+
Sbjct 62 INERSFICNYKQHWFTIRKFGKHWFNLNSLLAGPELISDTCLANFLARLQQQAYSVFVVK 121
> Hs13518013
Length=306
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 59/102 (57%), Gaps = 5/102 (4%)
Query 36 SGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVD----FGYILNHADHWLALR 91
SGN+ G+F++ V+ L+ + I N E++ +D +I N+ +HW +R
Sbjct 11 SGNMDDSGFFSIQVISNALKVWGLELIL-FNSPEYQRLRIDPINERSFICNYKEHWFTVR 69
Query 92 RVHGTWYNLDSMKPAPTVISSFHLEAFLSSLKGQGYTIFIVR 133
++ W+NL+S+ P +IS +L FL+ L+ +GY+IF+V+
Sbjct 70 KLGKQWFNLNSLLTGPELISDTYLALFLAQLQQEGYSIFVVK 111
> CE09760
Length=317
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 21/140 (15%)
Query 2 ELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHC 61
+L + +++D+ E+Q++ S N+ GYF++ VL + L +
Sbjct 38 DLRDLAIQMDKMEQQILGNANPTPG--------RSENMNESGYFSIQVLEKALETFSLKL 89
Query 62 ICQRNVGEHEEAGVDF--------GYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSF 113
N A VD+ YI N +HW LR+ W+ L+S+ P ++S
Sbjct 90 TNIEN-----PAMVDYKNNPLTARAYICNLREHWFVLRKFGNQWFELNSVNRGPKLLSDT 144
Query 114 HLEAFLSSLKGQGYTIFIVR 133
++ FL + +GY+IF+V+
Sbjct 145 YVSMFLHQVSSEGYSIFVVQ 164
> 7290691
Length=221
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 43/94 (45%), Gaps = 25/94 (26%)
Query 55 RRKRIHCICQRNVGEHEEAGVDFGYILN---------------HADHWLALRRVHGTWYN 99
RR+ HC+ V FG+ILN H HWLALRR++G++YN
Sbjct 115 RRRDPHCL---------NLSVIFGFILNVPAQMSLGYYIPLPFHMRHWLALRRLNGSYYN 165
Query 100 LDSMKPAPTVI-SSFHLEAFLSSLKGQGYTIFIV 132
LDS P + + FL++ + +F+V
Sbjct 166 LDSKLREPKCLGTEQQFLEFLATQLQMDHELFLV 199
> Hs19923879
Length=188
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 86 HWLALRRVHGTWYNLDSMKPAPTVISSFH-LEAFLSSLKGQG 126
HW+ALR+V G +YNLDS AP + + AFL++ QG
Sbjct 125 HWVALRQVDGVYYNLDSKLRAPEALGDEDGVRAFLAAALAQG 166
> At2g29640
Length=360
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 15/75 (20%)
Query 43 GYFNVSVLIECLRRKRIHCIC-QRNVG----EHEEAGVDFGYILN----------HADHW 87
G ++V+V+I L K + + +G + ++A G +LN + HW
Sbjct 71 GNYDVNVMITALEGKGKSVVWHDKRIGASSIDLDDADTLMGIVLNVPVKRYGGLWRSRHW 130
Query 88 LALRRVHGTWYNLDS 102
+ +R+++G WYNLDS
Sbjct 131 VVVRKINGVWYNLDS 145
> Hs7661888
Length=202
Score = 35.4 bits (80), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 20/112 (17%)
Query 42 DGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN--------------HA 84
+G ++V+V++ L+ K + +R+VG V G+I+N
Sbjct 79 NGNYDVNVIMAALQTKGYEAVWWDKRRDVGVIALTNV-MGFIMNLPSSLCWGPLKLPLKR 137
Query 85 DHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIVRP 134
HW+ +R V G +YNLDS P I L FL L+G+ + +V P
Sbjct 138 QHWICVREVGGAYYNLDSKLKMPEWIGGESELRKFLKHHLRGKNCELLLVVP 189
> At5g48080
Length=227
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 5/52 (9%)
Query 85 DHWLALRRVHGTWYNLDSMKPAPTVISS---FHLEAFLSSLKGQGYTIFIVR 133
D W +L++VHG Y+ ++MK + S F L + LSSL+ Y IF+VR
Sbjct 130 DVWSSLQKVHG--YDWNTMKRDEDLTKSKVCFFLWSLLSSLERIVYYIFLVR 179
> At3g54800
Length=709
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query 13 KERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEE 72
KE + G H D+ A +A G + ++ FN + + LR + C + NV EH +
Sbjct 193 KEAKDWDSRGRHWDDHPAIMAVGVIDGTSEDIFNTLMSLGPLRSEWDFCFYKGNVVEHLD 252
Query 73 AGVDFGYILNHADHWLA 89
D ++ ++D WL
Sbjct 253 GHTDIIHLQLYSD-WLP 268
> Hs13236559
Length=263
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 33 AEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVD 76
AE +AA GY NV L ++ +C+R G+ EE GVD
Sbjct 73 AEREALLAALGYKNVKKQPTGLSKEDFAEVCKREGGDPEEKGVD 116
> At1g49190
Length=608
Score = 28.9 bits (63), Expect = 3.4, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 46 NVSVLIECLRRKRIHCICQRNVGEH 70
N VL+ECL+ RI I + NV H
Sbjct 443 NPKVLVECLQEMRIEGITRSNVASH 467
> 7293885
Length=735
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 4/49 (8%)
Query 17 LMAEGGTHSKDYQAFLAEGSGNV--AADGYFNVSVLIECLRRKRIHCIC 63
+M+E G + YQ+++ E S + D YF V + RK IHC C
Sbjct 239 VMSETG--CRHYQSYVKEHSYDTFRVIDAYFAACVNRDARERKAIHCNC 285
> CE29383
Length=759
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 70 HEEAGVDFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVI 110
H+E + F + NH D L L++ G + M+ P V+
Sbjct 684 HDEILLFFASLANHMDATLVLQKARGLLFEFTRMEKIPCVL 724
Lambda K H
0.322 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40