bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6168_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
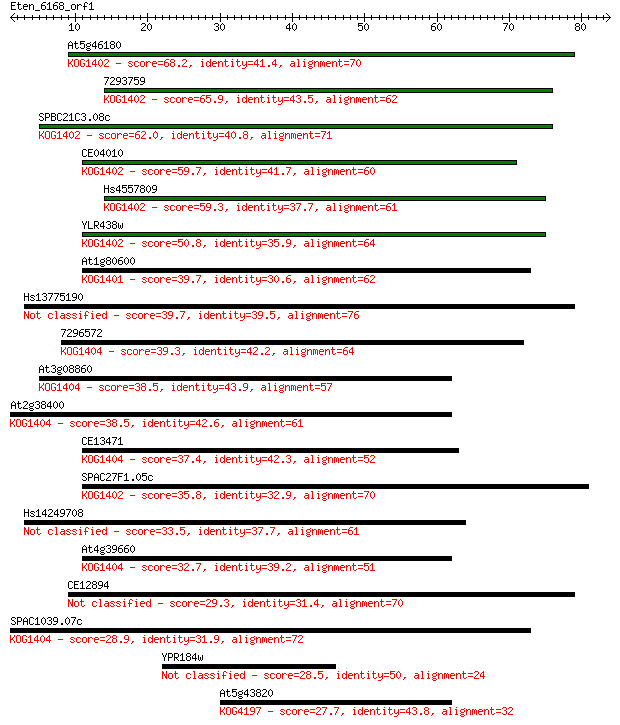
At5g46180 68.2 4e-12
7293759 65.9 2e-11
SPBC21C3.08c 62.0 2e-10
CE04010 59.7 1e-09
Hs4557809 59.3 2e-09
YLR438w 50.8 6e-07
At1g80600 39.7 0.001
Hs13775190 39.7 0.002
7296572 39.3 0.002
At3g08860 38.5 0.003
At2g38400 38.5 0.003
CE13471 37.4 0.006
SPAC27F1.05c 35.8 0.021
Hs14249708 33.5 0.10
At4g39660 32.7 0.18
CE12894 29.3 2.0
SPAC1039.07c 28.9 2.4
YPR184w 28.5 3.1
At5g43820 27.7 5.5
> At5g46180
Length=475
Score = 68.2 bits (165), Expect = 4e-12, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 9 EWIRDIRGRGLFNAVEINGSS---GEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEA 65
++I+++RGRGLFNA+E N S A +C+ LK GVL +PT ++R PPL I
Sbjct 371 KYIKEVRGRGLFNAIEFNSESLSPVSAYDICLSLKERGVLAKPTHNTIVRLTPPLSISSD 430
Query 66 QMKDALQRIQDVF 78
+++D + + DV
Sbjct 431 ELRDGSEALHDVL 443
> 7293759
Length=431
Score = 65.9 bits (159), Expect = 2e-11, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 45/62 (72%), Gaps = 1/62 (1%)
Query 14 IRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDALQR 73
+RG+GL NA+ IN +A ++C+ LK G+L +PT G+++RF PPL I+E QM++++
Sbjct 365 VRGKGLLNAIVIN-QKFDAWEVCLRLKENGLLAKPTHGDIIRFAPPLVINETQMRESIDI 423
Query 74 IQ 75
I+
Sbjct 424 IR 425
> SPBC21C3.08c
Length=438
Score = 62.0 bits (149), Expect = 2e-10, Method: Composition-based stats.
Identities = 29/74 (39%), Positives = 45/74 (60%), Gaps = 3/74 (4%)
Query 5 KCNLEWIRDIRGRGLFNAVEINGSSGE---AMQLCIELKNEGVLCRPTRGNVLRFLPPLC 61
+C ++ +RGRGL NAV I+ S A LC+ +++ GVL +PT GN++RF PPL
Sbjct 345 ECKSPIVQKVRGRGLLNAVVIDESKTNGRTAWDLCLIMRSRGVLAKPTHGNIIRFSPPLV 404
Query 62 IDEAQMKDALQRIQ 75
I E + ++ I+
Sbjct 405 ITEEDLMKGIEVIK 418
> CE04010
Length=422
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 43/60 (71%), Gaps = 1/60 (1%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDA 70
+ +RG+GLF A+ IN +A ++C++LK G+L + T G+++RF PPLCI++ Q++ A
Sbjct 349 VSTVRGKGLFCAIVINKKY-DAWKVCLKLKENGLLAKNTHGDIIRFAPPLCINKEQVEQA 407
> Hs4557809
Length=439
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 45/62 (72%), Gaps = 1/62 (1%)
Query 14 IRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDALQ 72
+RG+GL NA+ I + +A ++C+ L++ G+L +PT G+++RF PPL I E +++++++
Sbjct 371 VRGKGLLNAIVIKETKDWDAWKVCLRLRDNGLLAKPTHGDIIRFAPPLVIKEDELRESIE 430
Query 73 RI 74
I
Sbjct 431 II 432
> YLR438w
Length=424
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 11 IRDIRGRGLFNAVEINGSSGE---AMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQM 67
I ++RG GL A+ I+ S A LC+ +K+ G+L +PT +++R PPL I E +
Sbjct 351 ISEVRGMGLLTAIVIDPSKANGKTAWDLCLLMKDHGLLAKPTHDHIIRLAPPLVISEEDL 410
Query 68 KDALQRI 74
+ ++ I
Sbjct 411 QTGVETI 417
> At1g80600
Length=457
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 38/63 (60%), Gaps = 4/63 (6%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPT-RGNVLRFLPPLCIDEAQMKD 69
++++RG GL VE++ + + C ++ G+L +GNV+R +PPL I E +++
Sbjct 388 VKEVRGEGLIIGVELDVPASSLVDAC---RDSGLLILTAGKGNVVRIVPPLVISEEEIER 444
Query 70 ALQ 72
A++
Sbjct 445 AVE 447
> Hs13775190
Length=493
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 14/89 (15%)
Query 3 KEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGN 52
K+K I DIRG GLF +++ ++ EA + ++K + VL P R N
Sbjct 343 KQKAKHTLIGDIRGIGLFIGIDLVKDHLKRTPATAEAQHIIYKMKEKRVLLSADGPHR-N 401
Query 53 VLRFLPPLCIDEAQMK---DALQRIQDVF 78
VL+ PP+C E K D L RI V
Sbjct 402 VLKIKPPMCFTEEDAKFMVDQLDRILTVL 430
> 7296572
Length=470
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 37/74 (50%), Gaps = 10/74 (13%)
Query 8 LEWIRDIRGRGLFNAVEINGSSGEAMQLCI--------ELKNEGVLCRP--TRGNVLRFL 57
E I D+RG+GL VE+ G+ + L + K++GVL GNVLR
Sbjct 382 FEIIGDVRGKGLMIGVELVGNREKRTPLATPHVLDIWEKCKDQGVLLGRGGLHGNVLRIK 441
Query 58 PPLCIDEAQMKDAL 71
PP+CI A K A+
Sbjct 442 PPMCITLADAKFAV 455
> At3g08860
Length=481
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 30/66 (45%), Gaps = 9/66 (13%)
Query 5 KCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRPT--RGNVLR 55
K E I D+RGRGL VE + E + L ++K GVL GNV R
Sbjct 396 KNKYELIGDVRGRGLMLGVEFVKDRDLKTPAKAETLHLMDQMKEMGVLVGKGGFYGNVFR 455
Query 56 FLPPLC 61
PPLC
Sbjct 456 ITPPLC 461
> At2g38400
Length=477
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 11/70 (15%)
Query 1 KMKEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRPTR--G 51
++KEK E I D+RGRGL VE+ ++ E + + ++K GVL G
Sbjct 390 QLKEK--HEIIGDVRGRGLMLGVELVSDRKLKTPATAETLHIMDQMKELGVLIGKGGYFG 447
Query 52 NVLRFLPPLC 61
NV R PPLC
Sbjct 448 NVFRITPPLC 457
> CE13471
Length=506
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQ------LCIELKNEGVLCRP--TRGNVLRFLPPLCI 62
I D+RG+GL VE+ G+ + + + KN+G+L GNVLR PP+CI
Sbjct 389 IGDVRGKGLMIGVELIDEQGKPLAAAKTAAIFEDTKNQGLLIGKGGIHGNVLRIKPPMCI 448
> SPAC27F1.05c
Length=484
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 12/82 (14%)
Query 11 IRDIRGRGLFNAVE-------INGSSGE--AMQLCIELKNE---GVLCRPTRGNVLRFLP 58
++D+RGRG+ +E + GE A + +L + V C +V RFLP
Sbjct 389 MKDVRGRGMIVGIEFYPIPESVQEEFGEYYATPIVNDLADTYHVQVYCSLNNPSVFRFLP 448
Query 59 PLCIDEAQMKDALQRIQDVFCR 80
PL I EA + + L ++ +
Sbjct 449 PLTIPEADLDEGLSAVESAVAK 470
> Hs14249708
Length=175
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 11/71 (15%)
Query 3 KEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGN 52
++K + D+RG GLF V++ ++ EA L LK VL P R N
Sbjct 74 QQKIKHPIVGDVRGVGLFIGVDLIKDEATRTPATEEAAYLVSRLKENYVLLSTDGPGR-N 132
Query 53 VLRFLPPLCID 63
+L+F PP+C
Sbjct 133 ILKFKPPMCFS 143
> At4g39660
Length=447
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 28/60 (46%), Gaps = 9/60 (15%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRP--TRGNVLRFLPPLC 61
I D+RGRGL +E+ + E L +L+ G+L GNV R PP+C
Sbjct 365 IGDVRGRGLMVGIELVSDRKDKTPAKAETSVLFEQLRELGILVGKGGLHGNVFRIKPPMC 424
> CE12894
Length=467
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 10/80 (12%)
Query 9 EWIRDIRGRGLFNAVEI---NGSSGEAMQLCIEL-----KNEGVLCRPT--RGNVLRFLP 58
E I DIRG GLF +++ + +L I K+ G+L N+L+ P
Sbjct 383 ECIGDIRGVGLFWGIDLVKDRNTREPDQKLAIATILALRKSYGILLNADGPHTNILKIKP 442
Query 59 PLCIDEAQMKDALQRIQDVF 78
PLC +E + + + + V
Sbjct 443 PLCFNENNILETVTALDQVL 462
> SPAC1039.07c
Length=448
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 38/86 (44%), Gaps = 19/86 (22%)
Query 1 KMKEKCNLEWIRDIRGRGLFNAVEINGSSGEAM----------QLCIELKNEGVLCRPTR 50
++K+K L I D+RG GL +EI + + C+EL G+ C
Sbjct 351 RLKDKHPL--IVDVRGLGLLQGIEIASCTDPSKPSDFLGTVIGDKCLEL---GMNCNIVH 405
Query 51 ----GNVLRFLPPLCIDEAQMKDALQ 72
G V R PPL + + ++ A++
Sbjct 406 LRGIGGVFRIAPPLTVTDEEIHKAIE 431
> YPR184w
Length=1536
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 22 AVEINGSSGEAMQLCIELKNEGVL 45
A+EING A++ IELKN+G+
Sbjct 1271 AIEINGLLKSALRFVIELKNKGLF 1294
> At5g43820
Length=680
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 20/32 (62%), Gaps = 2/32 (6%)
Query 30 GEAMQLCIELKNEGVLCRPTRGNVLRFLPPLC 61
+A+++ E+ + GVL PT G V FL PLC
Sbjct 377 SDALEIFEEMLSRGVL--PTTGLVTSFLKPLC 406
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40