bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6058_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
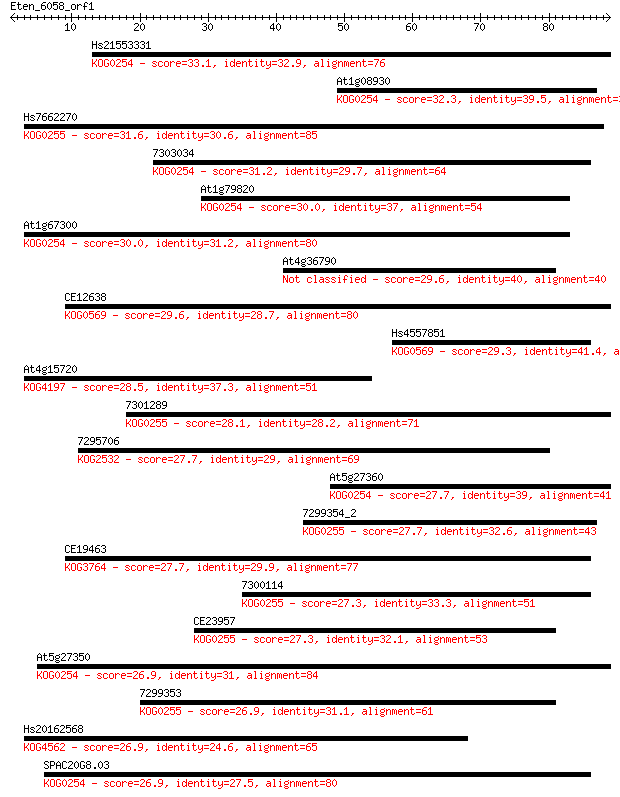
Hs21553331 33.1 0.11
At1g08930 32.3 0.20
Hs7662270 31.6 0.35
7303034 31.2 0.54
At1g79820 30.0 1.0
At1g67300 30.0 1.2
At4g36790 29.6 1.3
CE12638 29.6 1.6
Hs4557851 29.3 1.9
At4g15720 28.5 3.1
7301289 28.1 4.0
7295706 27.7 5.0
At5g27360 27.7 5.1
7299354_2 27.7 5.2
CE19463 27.7 5.9
7300114 27.3 6.3
CE23957 27.3 7.9
At5g27350 26.9 8.8
7299353 26.9 9.1
Hs20162568 26.9 9.3
SPAC20G8.03 26.9 9.6
> Hs21553331
Length=617
Score = 33.1 bits (74), Expect = 0.11, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 39/76 (51%), Gaps = 8/76 (10%)
Query 13 AAALGLLAVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAPGAAAGSLFGG 72
AA GLL G+ + + + AL Q++ +L +C E+ V+ L GA SL GG
Sbjct 47 AAVSGLLV---GYELGIISGALLQIKTLLAL-----SCHEQEMVVSSLVIGALLASLTGG 98
Query 73 FVADTVGRWRSLLWSS 88
+ D GR +++ SS
Sbjct 99 VLIDRYGRRTAIILSS 114
> At1g08930
Length=490
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 49 ACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRWRSLLW 86
+ AE F ++L G G++F G VAD +GR R++L+
Sbjct 87 SVAEYSMFGSILTLGGLIGAVFSGKVADVLGRKRTMLF 124
> Hs7662270
Length=683
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 36/85 (42%), Gaps = 5/85 (5%)
Query 3 CSSSSRLWWCAAALGLLAVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAP 62
C W LGL +A G V V + AL ++ L S +KG ++
Sbjct 102 CGHGRFQWILFFVLGLALMADGVEVFVVSFALPSAEKDMCLSSS-----KKGMLGMIVYL 156
Query 63 GAAAGSLFGGFVADTVGRWRSLLWS 87
G AG+ G +AD +GR R L S
Sbjct 157 GMMAGAFILGGLADKLGRKRVLSMS 181
> 7303034
Length=521
Score = 31.2 bits (69), Expect = 0.54, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Query 22 AAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRW 81
+ G + + A L QL+ + H ++ F +V A A G L GF+ D +GR
Sbjct 58 STGMTLAMPTATLHQLKD--TTEPVHLNDSQASWFASVNALSAPIGGLLSGFLLDRIGRK 115
Query 82 RSLL 85
+SL+
Sbjct 116 KSLI 119
> At1g79820
Length=472
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 3/54 (5%)
Query 29 VAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRWR 82
V L+ + G+ A +G V+ GA GSLF G VAD VGR R
Sbjct 46 VVNETLESISIDLGFSGNTIA---EGLVVSTCLGGAFIGSLFSGLVADGVGRRR 96
> At1g67300
Length=493
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 37/84 (44%), Gaps = 7/84 (8%)
Query 3 CSSSSRLWWCAAALGLLAVAA----GFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVA 58
+++ W C+ L+A + G+ + V L+ + G A +G V+
Sbjct 41 METTNPSWKCSLPHVLVATISSFLFGYHLGVVNEPLESISSDLGFSGDTLA---EGLVVS 97
Query 59 VLAPGAAAGSLFGGFVADTVGRWR 82
V GA GSLF G VAD GR R
Sbjct 98 VCLGGAFLGSLFSGGVADGFGRRR 121
> At4g36790
Length=489
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 41 FSLCGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVGR 80
F L G F + A + V A G A G+L GG +AD + R
Sbjct 286 FELIG--FDHNQTAALLGVFATGGAIGTLMGGIIADKMSR 323
> CE12638
Length=576
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 45/89 (50%), Gaps = 9/89 (10%)
Query 9 LWWCAAALGLLAVAAGFAV-CVAAAA---LQQLQQ-HFSLCGSHFACAEKGAFVAVLAPG 63
L++C +++ L + GF + C+ A + +++ HF L G + + +V
Sbjct 47 LFFCISSIALASFQDGFQIGCINAPGPLIIDWIKKCHFELFGEVLSQYQADFIWSVAVSM 106
Query 64 AAAGSLFG----GFVADTVGRWRSLLWSS 88
+ G +FG GF+AD GR +LL+++
Sbjct 107 FSVGGMFGSFCSGFLADKFGRKSTLLYNN 135
> Hs4557851
Length=524
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 57 VAVLAPGAAAGSLFGGFVADTVGRWRSLL 85
V+ A G S FGG++ DT+GR +++L
Sbjct 101 VSSFAVGGMTASFFGGWLGDTLGRIKAML 129
> At4g15720
Length=602
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 3 CSSSSRLWWCAAALGLLAVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEK 53
CSS RL W A GL+ + V A +L + ++ CGS +CAEK
Sbjct 246 CSSLGRLQWGKVAHGLVTRGGYESNTVVATSLLDM---YAKCGS-LSCAEK 292
> 7301289
Length=583
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 33/73 (45%), Gaps = 2/73 (2%)
Query 18 LLAVAAGFAVCVAAAALQQLQQHF--SLCGSHFACAEKGAFVAVLAPGAAAGSLFGGFVA 75
+L + +G + AA Q + S C AEKG +A G + G+++
Sbjct 332 VLLLVSGLLTITSVAAQQAMSIIVIASQCEFETTQAEKGVMMAASVTGIFLSTYIWGYIS 391
Query 76 DTVGRWRSLLWSS 88
D +GR R LL+ +
Sbjct 392 DDIGRRRVLLYGN 404
> 7295706
Length=439
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 32/75 (42%), Gaps = 6/75 (8%)
Query 11 WCAAALGLLAVAAGFAVCVAAAALQ--QLQQHFSLCGSHFACAEKGAFVAVLAPGAAAGS 68
W A ++ A+ A+ + ALQ SL + E+ +F +L G+A G+
Sbjct 115 WTAGSIKSYAIPFIVAIRILNGALQGVHFPSMISLTSQNLCPNERSSFFGLLTAGSALGT 174
Query 69 LF----GGFVADTVG 79
L G F+ D G
Sbjct 175 LLTGIMGSFLLDYFG 189
> At5g27360
Length=464
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Query 48 FACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRWRSLLWSS 88
+ A+ AF ++ GAA G+LF G +A +GR R +W S
Sbjct 55 LSIAQFSAFASLSTLGAAIGALFSGKMAIILGR-RKTMWVS 94
> 7299354_2
Length=508
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 20/43 (46%), Gaps = 0/43 (0%)
Query 44 CGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRWRSLLW 86
C +KG AV G ++ G++ADT GR + L W
Sbjct 33 CDLRLTLEDKGVLNAVAYAGMTISAIAWGYLADTKGRKKILYW 75
> CE19463
Length=626
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 12/84 (14%)
Query 9 LWWCAAALGLLAVAAG------FAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAP 62
LW +L +L +AA F +C+ + +F G C G F A
Sbjct 525 LWVIGISLAVLGLAASALYIPCFQMCLDEVKDKGFDDNFHTYG----CVS-GIFQAAFGF 579
Query 63 GAAAGSLFGGFVADTVG-RWRSLL 85
G+ G FG V + VG RW + +
Sbjct 580 GSFIGPTFGSVVVERVGFRWTTTM 603
> 7300114
Length=670
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Query 35 QQLQQHFSL-CGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVGRWRSLL 85
+ L Q F L CG + F+ GAAAG++ G+++D GR +L+
Sbjct 262 KTLVQEFDLVCGRDILSLVETCFLV----GAAAGAVLSGWISDRFGRRHTLM 309
> CE23957
Length=591
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 30/57 (52%), Gaps = 4/57 (7%)
Query 28 CVAAAALQQLQQHFSLCGS-HFAC--AEKGAFVAVLAPGAA-AGSLFGGFVADTVGR 80
C A ++Q + FS+ G C ++K ++ V+ G++ GS+ GG + D GR
Sbjct 105 CTNLAPVKQNLEFFSIVGQFQLICDDSDKVEYIEVIMAGSSLIGSIIGGHMGDHFGR 161
> At5g27350
Length=469
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 39/88 (44%), Gaps = 10/88 (11%)
Query 5 SSSRLWWCAAALGLLAVAAGFAVCVA----AAALQQLQQHFSLCGSHFACAEKGAFVAVL 60
S R+ C +AV F+ VA + A + + L + F+ AF +
Sbjct 21 SECRITACVILSTFVAVCGSFSFGVATGYTSGAETGVMKDLDLSIAQFS-----AFGSFA 75
Query 61 APGAAAGSLFGGFVADTVGRWRSLLWSS 88
GAA G+LF G +A +GR R +W S
Sbjct 76 TLGAAIGALFCGNLAMVIGR-RGTMWVS 102
> 7299353
Length=488
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 20 AVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAPGAAAGSLFGGFVADTVG 79
AV A+ +A+AL + + C + +KG AV G ++ GF+ADT+G
Sbjct 35 AVPCLTAMVFSASALSYVMPS-AECDLDLSIVDKGMLHAVTYAGMIISAVPWGFIADTIG 93
Query 80 R 80
R
Sbjct 94 R 94
> Hs20162568
Length=643
Score = 26.9 bits (58), Expect = 9.3, Method: Composition-based stats.
Identities = 16/65 (24%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 3 CSSSSRLWWCAAALGLLAVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAP 62
C+S +W A+G + AGFA ++ AL + + + C + +G+ + + P
Sbjct 289 CASEEVIWEVLNAIGPWSALAGFADVLSRLALWESEGPEAFCEESGLRSAEGSVLDLANP 348
Query 63 GAAAG 67
AG
Sbjct 349 QGLAG 353
> SPAC20G8.03
Length=557
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 3/80 (3%)
Query 6 SSRLWWCAAALGLLAVAAGFAVCVAAAALQQLQQHFSLCGSHFACAEKGAFVAVLAPGAA 65
SS +W +A G+ + G+ V + AL L G + +K + + A
Sbjct 78 SSWIWVLSAVAGISGLLFGYDTGVISGALAVLGSDL---GHVLSSGQKELITSATSFAAL 134
Query 66 AGSLFGGFVADTVGRWRSLL 85
+ G++AD VGR R LL
Sbjct 135 ISATTSGWLADWVGRKRLLL 154
Lambda K H
0.328 0.132 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40