bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
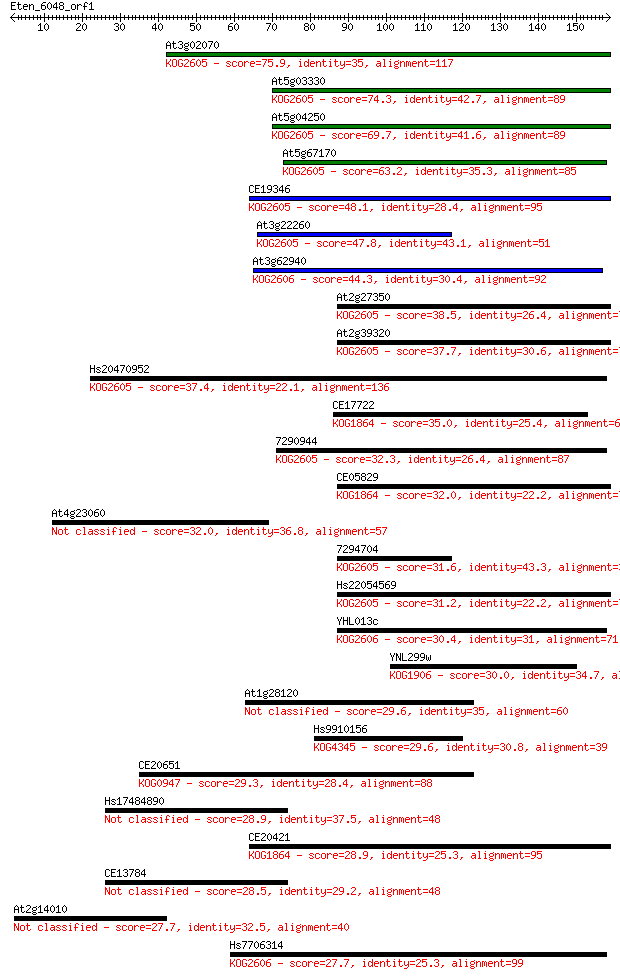
Query= Eten_6048_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
At3g02070 75.9 3e-14
At5g03330 74.3 9e-14
At5g04250 69.7 2e-12
At5g67170 63.2 2e-10
CE19346 48.1 8e-06
At3g22260 47.8 9e-06
At3g62940 44.3 9e-05
At2g27350 38.5 0.005
At2g39320 37.7 0.010
Hs20470952 37.4 0.011
CE17722 35.0 0.062
7290944 32.3 0.34
CE05829 32.0 0.47
At4g23060 32.0 0.52
7294704 31.6 0.73
Hs22054569 31.2 0.89
YHL013c 30.4 1.3
YNL299w 30.0 1.7
At1g28120 29.6 2.2
Hs9910156 29.6 2.5
CE20651 29.3 3.6
Hs17484890 28.9 4.2
CE20421 28.9 4.3
CE13784 28.5 6.3
At2g14010 27.7 8.4
Hs7706314 27.7 9.6
> At3g02070
Length=219
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 62/118 (52%), Gaps = 4/118 (3%)
Query 42 SSTSPRAAARAHVSYPKNITAWETESSLLEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFG 101
S+ +P Y N+ + L QRL+ G + GDGNCQFR+ S L+
Sbjct 42 SNLAPVPHVPRINCYIPNLNDATLDHQRLLQRLNVYGLCELKVSGDGNCQFRALSDQLYR 101
Query 102 KEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPL-FDRYLRDMERSGTWGDELSLRAVAD 158
+ H+ VRR V Q+++CR YE + P+ + RY + M + G WGD ++L+A AD
Sbjct 102 SPEYHKQVRREVVKQLKECRSMYESY---VPMKYKRYYKKMGKFGEWGDHITLQAAAD 156
> At5g03330
Length=356
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Query 70 LEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFD 129
L RL V GDGNCQFR+ + L+ D H+HVRR V Q++ Y+ +
Sbjct 204 LRNRLEMFDFTEVKVPGDGNCQFRALADQLYKTADRHKHVRRQIVKQLKSRPDSYQGY-- 261
Query 130 GAPL-FDRYLRDMERSGTWGDELSLRAVAD 158
P+ F YLR M RSG WGD ++L+A AD
Sbjct 262 -VPMDFSDYLRKMSRSGEWGDHVTLQAAAD 290
> At5g04250
Length=395
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 50/89 (56%), Gaps = 2/89 (2%)
Query 70 LEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFD 129
L QRL G GDGNCQFRS S L+ + H VR V Q+ R+ YE +
Sbjct 195 LFQRLQLYGLVENKIEGDGNCQFRSLSDQLYRSPEHHNFVREQVVNQLAYNREIYEGYVP 254
Query 130 GAPLFDRYLRDMERSGTWGDELSLRAVAD 158
A ++ YL+ M+R+G WGD ++L+A AD
Sbjct 255 MA--YNDYLKAMKRNGEWGDHVTLQAAAD 281
> At5g67170
Length=382
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 45/85 (52%), Gaps = 0/85 (0%)
Query 73 RLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAP 132
+L LG + + DGNC FR+ + L G EDEH R M V + K R+ +E F +
Sbjct 38 QLDALGLKIIQVTADGNCFFRAIADQLEGNEDEHNKYRNMIVLYIVKNREMFEPFIEDDV 97
Query 133 LFDRYLRDMERSGTWGDELSLRAVA 157
F+ Y + M+ GTW + L+A +
Sbjct 98 PFEDYCKTMDDDGTWAGNMELQAAS 122
> CE19346
Length=442
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 47/95 (49%), Gaps = 5/95 (5%)
Query 64 ETESSLLEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKD 123
ETE S RL+ G VGDG C FRS + ++G ++ H +RR+ + M R
Sbjct 124 ETEFS---NRLAARGLIIKEMVGDGACMFRSIAEQIYGDQEMHGQIRRLCMDYMSNNRDH 180
Query 124 YEVFFDGAPLFDRYLRDMERSGTWGDELSLRAVAD 158
++ F F+ Y++ G+ + L+A+++
Sbjct 181 FKEFITEN--FENYIQRKREENVHGNHVELQAISE 213
> At3g22260
Length=205
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 66 ESSLLEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQ 116
+ LL RL+ G + GDGNCQFR+ + LF D H+HVR+ V Q
Sbjct 88 DHELLSGRLATYGLAELQMEGDGNCQFRALADQLFRNADYHKHVRKHVVKQ 138
> At3g62940
Length=332
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 16/108 (14%)
Query 65 TESSLLEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKED-----EHRHVRRMAVAQMRK 119
E++ LE++L LG DG+C +R+ L + ++++R MA + MR+
Sbjct 167 VENAKLEKKLKPLGLTVSEIKPDGHCLYRAVENQLANRSGGASPYTYQNLREMAASYMRE 226
Query 120 CRKDYEVFF-----------DGAPLFDRYLRDMERSGTWGDELSLRAV 156
+ D+ FF F++Y R++E + WG +L L A+
Sbjct 227 HKTDFLPFFLSETEGDSNSGSAEERFEKYCREVESTAAWGSQLELGAL 274
> At2g27350
Length=522
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 2/72 (2%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLRDMERSGT 146
DGNC FR+ + ++G + + R+M + M + R + F F YL+ R
Sbjct 270 DGNCLFRAVADQVYGDSEAYDLARQMCMDYMEQERDHFSQFITEG--FTSYLKRKRRDKV 327
Query 147 WGDELSLRAVAD 158
+G+ + ++A+A+
Sbjct 328 YGNNVEIQALAE 339
> At2g39320
Length=189
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 30/72 (41%), Gaps = 21/72 (29%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLRDMERSGT 146
DGNCQFR+ + L+ D H VR+ V Q + +
Sbjct 5 DGNCQFRALADQLYQNSDCHELVRQEIVKQN---------------------MSLSTNSQ 43
Query 147 WGDELSLRAVAD 158
WGDE++LR AD
Sbjct 44 WGDEVTLRVAAD 55
> Hs20470952
Length=470
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 30/142 (21%), Positives = 56/142 (39%), Gaps = 15/142 (10%)
Query 22 KKSDREEEARPTTVLTTTSNSSTSPRAAARAHVSYPKN------ITAWETESSLLEQRLS 75
++ D E EA P + +S+ P +R S P++ + E + L QR
Sbjct 242 RRPDPEAEAPPAGSIEAAPSSAAEPVIVSR---SDPRDEKLALYLAEVEKQDKYLRQRNK 298
Query 76 FLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFD 135
+ + DGNC +R+ S +++G + HR +R V + + +G
Sbjct 299 Y----RFHIIPDGNCLYRAVSKTVYGDQSLHRELREQTVHYIADHLDHFSPLIEGD--VG 352
Query 136 RYLRDMERSGTWGDELSLRAVA 157
++ + G W L A+
Sbjct 353 EFIIAAAQDGAWAGYPELLAMG 374
> CE17722
Length=532
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 86 GDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLRDMERSG 145
DGNC +R+ S+ L G E H R ++K ++ + ++++Y+ ++ R G
Sbjct 330 ADGNCFYRAVSWWLTGVESHHMIFREAVGKHLKKNEPKFKKYCHDE-IYEKYVDNVMREG 388
Query 146 TWGDELS 152
W S
Sbjct 389 VWASTYS 395
> 7290944
Length=811
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 39/87 (44%), Gaps = 2/87 (2%)
Query 71 EQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDG 130
+Q L G + D + FR + ++ + H +R V M R+ +E G
Sbjct 21 DQYLESRGLYRKHTARDASSLFRVIAEQMYDTQMLHYEIRLECVRFMTLKRRIFEKEIPG 80
Query 131 APLFDRYLRDMERSGTWGDELSLRAVA 157
FD Y++DM + T+G LRA++
Sbjct 81 D--FDSYMQDMSKPKTYGTMTELRAMS 105
> CE05829
Length=781
Score = 32.0 bits (71), Expect = 0.47, Method: Composition-based stats.
Identities = 16/72 (22%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLRDMERSGT 146
DGNC +R+ S+ + G + H R ++K ++ + ++++Y+ + R G
Sbjct 563 DGNCFYRAISWWITGVQSHHMIFREAVGKHLKKNELMFKKYCHNE-IYEKYVENAMRDGV 621
Query 147 WGDELSLRAVAD 158
W + A+A+
Sbjct 622 WATTCEIFAMAN 633
> At4g23060
Length=543
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 27/61 (44%), Gaps = 4/61 (6%)
Query 12 ASFFRRLWSFKKSDREEEARPTTVLTTTSNSSTSPRAAARAHVSYP----KNITAWETES 67
+S +R WSF KS RE+E+ P + T + S H S P K WE E
Sbjct 34 SSNLKRRWSFVKSKREKESTPINQVPHTPSLPNSTPPPPSHHQSSPRRRRKQKPMWEDEG 93
Query 68 S 68
S
Sbjct 94 S 94
> 7294704
Length=649
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQ 116
DG C FRS S ++G E+ H H R Q
Sbjct 224 DGACLFRSISLQIYGDEEMHDHENREYFGQ 253
> Hs22054569
Length=591
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 16/72 (22%), Positives = 35/72 (48%), Gaps = 2/72 (2%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLRDMERSGT 146
DG C FR+ + ++G +D H VR+ + + K + + F Y+ ++
Sbjct 197 DGACLFRAVADQVYGDQDMHEVVRKHCMDYLMKNADYFSNYVTED--FTTYINRKRKNNC 254
Query 147 WGDELSLRAVAD 158
G+ + ++A+A+
Sbjct 255 HGNHIEMQAMAE 266
> YHL013c
Length=307
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 13/84 (15%)
Query 87 DGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMR--------KCRKDY-EVFFDGAPL---- 133
DG+C F S L + D + + M V ++R + R D+ FD +
Sbjct 175 DGHCLFASILDQLKLRHDPKKLDQDMDVMKLRWLSCNYVQEHRDDFIPYLFDEETMKMKD 234
Query 134 FDRYLRDMERSGTWGDELSLRAVA 157
D Y ++ME + WG E+ + A++
Sbjct 235 IDEYTKEMEHTAQWGGEIEILALS 258
> YNL299w
Length=625
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 3/52 (5%)
Query 101 GKEDEHRHVRRMAVAQMRKCRKDYEVFFDGAPLFDRYLR---DMERSGTWGD 149
GK E +H +RM + + +K EVF D F++Y D+E T+G+
Sbjct 13 GKRKEDKHTKRMRKSSFTRTQKMLEVFNDNRSHFNKYESLAIDVEDDDTFGN 64
> At1g28120
Length=306
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 7/66 (10%)
Query 63 WETESSLLEQRLSFLGCRTVT---SVGDGNCQFRSCSFSLFGKEDEHR---HVRRMAVAQ 116
+++ S +L +++ L + + + GDGNC FRS FS E + V R+ V
Sbjct 62 YQSGSPILLEKIKILDSQYIGIRRTRGDGNCFFRSFMFSYLEHILESQDRAEVDRIKV-N 120
Query 117 MRKCRK 122
+ KCRK
Sbjct 121 VEKCRK 126
> Hs9910156
Length=858
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 81 TVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRK 119
+ + GDGNC + S ++G D +R+ A M K
Sbjct 200 PLATTGDGNCLLHAASLGMWGFHDRDLMLRKALYALMEK 238
> CE20651
Length=1260
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 34/88 (38%), Gaps = 12/88 (13%)
Query 35 VLTTTSNSSTSPRAAARAHVSYPKNITAWETESSLLEQRLSFLGCRTVTSVGDGNCQFRS 94
V + ++ +T+ AAA PK+ +W TE + L F G R FR
Sbjct 867 VASADADETTNKNAAAIGFSKLPKSELSWLTEENSLLSTSKF-GTRGAAHCSTNIRSFRL 925
Query 95 CSFSLFGKEDEHRHVRRMAVAQMRKCRK 122
C L G VA M+KC K
Sbjct 926 CEIPLSG-----------IVAVMKKCAK 942
> Hs17484890
Length=1039
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 26 REEEARPTTVLTTTSNSSTSPRAAARAH-VSYPKNITAWETESSLLEQR 73
R+ RP +VLT ++ SS SP+ R H V +++ T SS QR
Sbjct 358 RQRPHRPDSVLTASTASSPSPQCRHRVHSVLNVRSVLTAPTASSPRPQR 406
> CE20421
Length=975
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 43/97 (44%), Gaps = 6/97 (6%)
Query 64 ETESSLLEQRLSFLGCRTVTSVGDGNCQFRSCSFSLFGKEDEHRHVRRMAVAQMRKCRKD 123
E E S++ ++ G R DGNC +R+ S+ L G + H+ +R +A R D
Sbjct 699 ENEKSVVMKKFDRPG-RVKGIESDGNCFYRAISWCLTGSQKYHKALR---IATANYLRND 754
Query 124 YEVFFDGAPLFDR--YLRDMERSGTWGDELSLRAVAD 158
+ D Y++ +E G W + + +A+
Sbjct 755 IAIVDKYCHKTDHKTYVQQVEGDGWWATNVEICVMAN 791
> CE13784
Length=691
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 14/56 (25%), Positives = 25/56 (44%), Gaps = 8/56 (14%)
Query 26 REEEARPTTVLTTTSNSSTSPRAAA--------RAHVSYPKNITAWETESSLLEQR 73
++E + P T T + +T+P+ A + H+ Y T + E L+E R
Sbjct 417 KDEPSYPLTTTTAVTEMNTTPKTIATSPTSKDPKLHLQYDNTTTTFTQEDKLIELR 472
> At2g14010
Length=833
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 5/45 (11%)
Query 2 QGDEKQLARGASFFRRLWSFK-----KSDREEEARPTTVLTTTSN 41
+ D+ ++ + RLWS K K DR+ + + T+V++ T+N
Sbjct 220 ENDDSEMEETQTEIERLWSLKLDIVWKVDRQSQTKVTSVMSGTAN 264
> Hs7706314
Length=289
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 25/117 (21%), Positives = 44/117 (37%), Gaps = 25/117 (21%)
Query 59 NIT-AWETESSLLEQRLSFLGCRTVTSVGDGNCQFRS---------CSFSLFGKEDEHRH 108
N+T A ES L Q L+ DG+C +++ C+ ++
Sbjct 126 NLTGARHMESEKLAQILAARQLEIKQIPSDGHCMYKAIEDQLKEKDCALTVVA------- 178
Query 109 VRRMAVAQMRKCRKDYEVFFDGAPL--------FDRYLRDMERSGTWGDELSLRAVA 157
+R M+ +D+ F F +Y D+ + WG +L LRA++
Sbjct 179 LRSQTAEYMQSHVEDFLPFLTNPNTGDMYTPEEFQKYCEDIVNTAAWGGQLELRALS 235
Lambda K H
0.320 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40