bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6028_orf1
Length=99
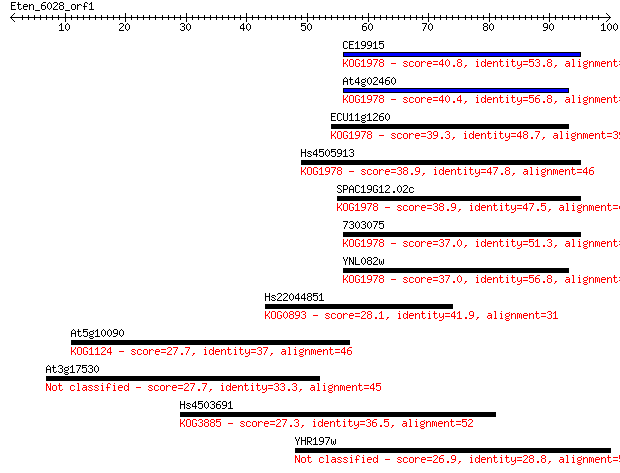
Score E
Sequences producing significant alignments: (Bits) Value
CE19915 40.8 6e-04
At4g02460 40.4 8e-04
ECU11g1260 39.3 0.002
Hs4505913 38.9 0.002
SPAC19G12.02c 38.9 0.002
7303075 37.0 0.009
YNL082w 37.0 0.009
Hs22044851 28.1 4.2
At5g10090 27.7 5.7
At3g17530 27.7 6.1
Hs4503691 27.3 6.9
YHR197w 26.9 9.3
> CE19915
Length=805
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 56 LFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSLPA 94
LFIVDQHASDEK FE L + QPL P L A
Sbjct 638 LFIVDQHASDEKYNFERLQSSAKLTKQPLFMPTALGFGA 676
> At4g02460
Length=779
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/37 (56%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 56 LFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSL 92
LFIVDQHA+DEK FE L QPL+ PL L L
Sbjct 580 LFIVDQHAADEKFNFEHLARSTVLNQQPLLQPLNLEL 616
> ECU11g1260
Length=630
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 54 TSLFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSL 92
T L VDQHA+DE R FE + F + Q +I+P+KL L
Sbjct 477 TYLVAVDQHAADEIRNFEDIKKTFCLKKQSVISPVKLDL 515
> Hs4505913
Length=862
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 49 IEEVVTSLFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSLPA 94
I ++ +FIVDQHA+DEK FE L + Q LIAP L+L A
Sbjct 688 ITKLNEDIFIVDQHATDEKYNFEMLQQHTVLQGQRLIAPQTLNLTA 733
> SPAC19G12.02c
Length=794
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 55 SLFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSLPA 94
+LFI+DQHASDEK +E L ++ Q L+ P +L L A
Sbjct 629 NLFIIDQHASDEKFNYEHLKSNLVINSQDLVLPKRLDLAA 668
> 7303075
Length=899
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 56 LFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSLPA 94
LFIVDQHA+DEK FE L Q L P L L A
Sbjct 720 LFIVDQHATDEKYNFETLQRTTQLEYQRLAVPQNLELTA 758
> YNL082w
Length=904
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 21/37 (56%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 56 LFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSL 92
LFIVDQHASDEK FE L A + Q LI P + L
Sbjct 728 LFIVDQHASDEKYNFETLQAVTVFKSQKLIIPQPVEL 764
> Hs22044851
Length=93
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 43 RKSCEEIEEVVTSLFIVDQHASDEKRIFEAL 73
+K I EVVT +I+D H +KR+ +AL
Sbjct 6 KKGRSAINEVVTEEYIIDIHKGFKKRVSQAL 36
> At5g10090
Length=594
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 11 PQITQAREYSNSTCNGKYAENYGSSSSSREVIRKSCEEIEEVVTSL 56
P T+AR + CN K N+ S+ E++RK E EEV+ L
Sbjct 536 PGYTKAR-LRRADCNAKLG-NWESAVGDYEILRKETPEDEEVIKGL 579
> At3g17530
Length=388
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 10/48 (20%)
Query 7 RCLFPQITQAREYSNSTCNGKYAENYGSSSS---SREVIRKSCEEIEE 51
RC+ P+I CN +YA YG+S S S +++R C +++
Sbjct 129 RCIKPRIFY-------RCNDRYALGYGNSKSSCHSYKILRSCCYYVDQ 169
> Hs4503691
Length=207
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 6/55 (10%)
Query 29 AENYGSSSSSRE-VIRKSCEE--IEEVVTSLFIVDQHASDEKRIFEALNADFHPR 80
E YGS +RE V R+ EE ++L+ +H+ E++ + ALN D PR
Sbjct 121 GELYGSKKLTRECVFREQFEENWYNTYASTLY---KHSDSERQYYVALNKDGSPR 172
> YHR197w
Length=763
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 48 EIEEVVTSLFIVDQHASDEKRIFEALNADFHPRMQPLIAPLKLSLPADLLAA 99
E+ +++ SL ++D+ +++++ +L DF+ + P +LSL AD+L A
Sbjct 272 ELYKLIKSLPVIDE-SNNKEEFLPSLKLDFNAPLTLWEIPQRLSLLADMLVA 322
Lambda K H
0.318 0.131 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40