bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5863_orf2
Length=68
Score E
Sequences producing significant alignments: (Bits) Value
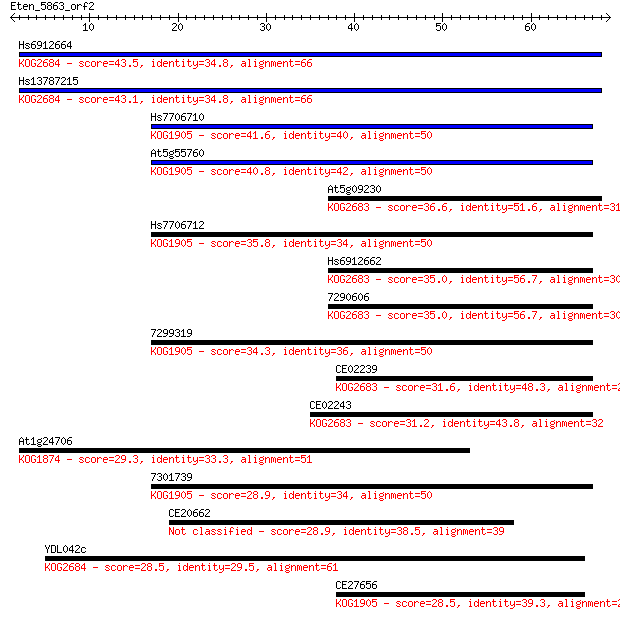
Hs6912664 43.5 9e-05
Hs13787215 43.1 1e-04
Hs7706710 41.6 4e-04
At5g55760 40.8 6e-04
At5g09230 36.6 0.010
Hs7706712 35.8 0.021
Hs6912662 35.0 0.031
7290606 35.0 0.033
7299319 34.3 0.061
CE02239 31.6 0.39
CE02243 31.2 0.48
At1g24706 29.3 1.9
7301739 28.9 2.3
CE20662 28.9 2.3
YDL042c 28.5 2.9
CE27656 28.5 3.6
> Hs6912664
Length=310
Score = 43.5 bits (101), Expect = 9e-05, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 37/73 (50%), Gaps = 9/73 (12%)
Query 2 WQRYDPAVYATIWGFWRQPSKIWQLLH---EFLNEREPEPNRAHLALAQLEA----LGIV 54
W+++ AT F PS++W+ H E + + EPN H A+A+ E G
Sbjct 77 WRKWQAQDLATPLAFAHNPSRVWEFYHYRREVMGSK--EPNAGHRAIAECETRLGKQGRR 134
Query 55 RAIVTQNVDNLHQ 67
++TQN+D LH+
Sbjct 135 VVVITQNIDELHR 147
> Hs13787215
Length=299
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 37/73 (50%), Gaps = 9/73 (12%)
Query 2 WQRYDPAVYATIWGFWRQPSKIWQLLH---EFLNEREPEPNRAHLALAQLEA----LGIV 54
W+++ AT F PS++W+ H E + + EPN H A+A+ E G
Sbjct 77 WRKWQAQDLATPLAFAHNPSRVWEFYHYRREVMGSK--EPNAGHRAIAECETRLGKQGRR 134
Query 55 RAIVTQNVDNLHQ 67
++TQN+D LH+
Sbjct 135 VVVITQNIDELHR 147
> Hs7706710
Length=355
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 31/56 (55%), Gaps = 6/56 (10%)
Query 17 WRQPSKIWQLLHEFLNER------EPEPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+R P +W + L + P + H+AL QLE +G++R +V+QNVD LH
Sbjct 64 FRGPHGVWTMEERGLAPKFDTTFESARPTQTHMALVQLERVGLLRFLVSQNVDGLH 119
> At5g55760
Length=473
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query 17 WRQPSKIWQLLHE-------FLNEREPEPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+R P IW L E L P+ H+AL +LE GI++ +++QNVD LH
Sbjct 64 FRGPKGIWTLQREGKDLPKASLPFHRAMPSMTHMALVELERAGILKFVISQNVDGLH 120
> At5g09230
Length=451
Score = 36.6 bits (83), Expect = 0.010, Method: Composition-based stats.
Identities = 16/31 (51%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 37 EPNRAHLALAQLEALGIVRAIVTQNVDNLHQ 67
+P AH ALA LE G + ++TQNVD LH
Sbjct 232 QPGPAHTALASLEKAGRINFMITQNVDRLHH 262
> Hs7706712
Length=400
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 30/55 (54%), Gaps = 5/55 (9%)
Query 17 WRQPSKIWQLLHE-----FLNEREPEPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+R P+ +W LL + + E EP H+++ +L +V+ +V+QN D LH
Sbjct 119 YRGPNGVWTLLQKGRSVSAADLSEAEPTLTHMSITRLHEQKLVQHVVSQNCDGLH 173
> Hs6912662
Length=314
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 37 EPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+PN AH AL+ E LG + +VTQNVD LH
Sbjct 120 QPNPAHWALSTWEKLGKLYWLVTQNVDALH 149
> 7290606
Length=204
Score = 35.0 bits (79), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 37 EPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+PN H ALA+ E V+A+VTQNVD LH
Sbjct 111 QPNATHHALARFEREERVQAVVTQNVDRLH 140
> 7299319
Length=317
Score = 34.3 bits (77), Expect = 0.061, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 10/58 (17%)
Query 17 WRQPSKIWQLLHEFLNER--------EPEPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+R P +W L E E+ E P + H+A+ L G V+ +++QN+D LH
Sbjct 64 FRGPKGVWTL--EEKGEKPDFNVSFDEARPTKTHMAIIALIESGYVQYVISQNIDGLH 119
> CE02239
Length=287
Score = 31.6 bits (70), Expect = 0.39, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 38 PNRAHLALAQLEALGIVRAIVTQNVDNLH 66
PN H AL++ EA ++TQNVD LH
Sbjct 94 PNFNHYALSKWEAANKFHWLITQNVDGLH 122
> CE02243
Length=287
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 35 EPEPNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+ PN H AL++ EA + ++TQNVD LH
Sbjct 91 QAAPNINHYALSKWEASDRFQWLITQNVDGLH 122
> At1g24706
Length=1705
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 21/51 (41%), Gaps = 6/51 (11%)
Query 2 WQRYDPAVYATIWGFWRQPSKIWQLLHEFLNEREPEPNRAHLALAQLEALG 52
W P +YAT WG LH N E E ++ H AL LE +
Sbjct 816 WNSLSPDLYATFWGL------TLYDLHVPRNRYESEISKQHTALKTLEEVA 860
> 7301739
Length=753
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Query 17 WRQPSKIWQLLHEFLNEREPE-----PNRAHLALAQLEALGIVRAIVTQNVDNLH 66
+R IW LL + + E + P H+AL +L ++ +V+QN D LH
Sbjct 125 YRGSQGIWTLLQKGQDIGEHDLSSANPTYTHMALYELHRRRLLHHVVSQNCDGLH 179
> CE20662
Length=367
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query 19 QPSKIWQLLHEFLNEREPEPNRAHLALAQ-----LEALGIVRAI 57
Q +WQL H F+ + EP + +H ++ Q L A G+V I
Sbjct 324 QNRNLWQLYHSFVIQVEPCKSSSHFSMFQTIATLLSAAGLVSKI 367
> YDL042c
Length=562
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 14/61 (22%)
Query 5 YDPAVYATIWGFWRQPSKIWQLLHEFLNEREPEPNRAHLALAQLEALGIVRAIVTQNVDN 64
+DP+V+ I P KI+ LH F+ L+ G + TQN+DN
Sbjct 303 HDPSVFYNIANMVLPPEKIYSPLHSFIK--------------MLQMKGKLLRNYTQNIDN 348
Query 65 L 65
L
Sbjct 349 L 349
> CE27656
Length=299
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 38 PNRAHLALAQLEALGIVRAIVTQNVDNL 65
P +H ++ L G ++ I+TQNVD L
Sbjct 101 PGVSHKSILALHKAGYIKTIITQNVDGL 128
Lambda K H
0.323 0.137 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1203543208
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40