bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
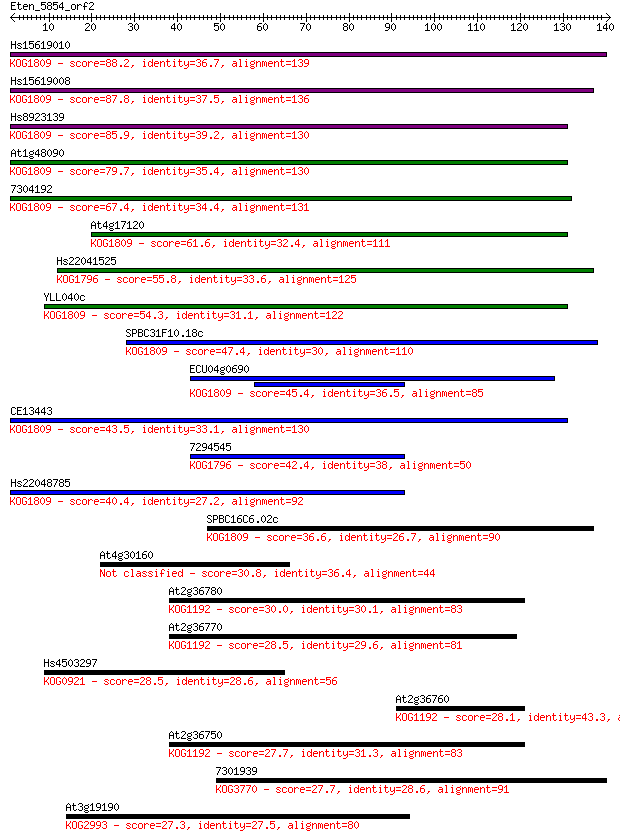
Query= Eten_5854_orf2
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
Hs15619010 88.2 5e-18
Hs15619008 87.8 7e-18
Hs8923139 85.9 2e-17
At1g48090 79.7 2e-15
7304192 67.4 9e-12
At4g17120 61.6 5e-10
Hs22041525 55.8 2e-08
YLL040c 54.3 7e-08
SPBC31F10.18c 47.4 9e-06
ECU04g0690 45.4 3e-05
CE13443 43.5 1e-04
7294545 42.4 3e-04
Hs22048785 40.4 0.001
SPBC16C6.02c 36.6 0.016
At4g30160 30.8 0.88
At2g36780 30.0 1.6
At2g36770 28.5 4.0
Hs4503297 28.5 4.1
At2g36760 28.1 5.4
At2g36750 27.7 7.0
7301939 27.7 7.1
At3g19190 27.3 9.1
> Hs15619010
Length=3174
Score = 88.2 bits (217), Expect = 5e-18, Method: Composition-based stats.
Identities = 51/142 (35%), Positives = 74/142 (52%), Gaps = 11/142 (7%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEG-VWSLTNIVTKPIEGAQREGV 59
++ T D +Y +R+ + A R+G GK + G V +T IVTKPI+GAQ+ G
Sbjct 2932 VAAMTMDEDYQQKRREAMNKQPAGFREGITRGGKGLVSGFVSGITGIVTKPIKGAQKGGA 2991
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIK--AEVSKPIGASKFRNERRRKPRMLG 117
GFFKG+GKG+VG++ +P + S +GIK E S+ E R PR
Sbjct 2992 AGFFKGVGKGLVGAVARPTGGIIDMASSTFQGIKRATETSEV--------ESLRPPRFFN 3043
Query 118 ELGEIRPYDETEATLRECLGLL 139
E G IRPY + T + L ++
Sbjct 3044 EDGVIRPYRLRDGTGNQMLQVM 3065
> Hs15619008
Length=3095
Score = 87.8 bits (216), Expect = 7e-18, Method: Composition-based stats.
Identities = 51/139 (36%), Positives = 72/139 (51%), Gaps = 11/139 (7%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEG-VWSLTNIVTKPIEGAQREGV 59
++ T D +Y +R+ + A R+G GK + G V +T IVTKPI+GAQ+ G
Sbjct 2932 VAAMTMDEDYQQKRREAMNKQPAGFREGITRGGKGLVSGFVSGITGIVTKPIKGAQKGGA 2991
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIK--AEVSKPIGASKFRNERRRKPRMLG 117
GFFKG+GKG+VG++ +P + S +GIK E S+ E R PR
Sbjct 2992 AGFFKGVGKGLVGAVARPTGGIIDMASSTFQGIKRATETSEV--------ESLRPPRFFN 3043
Query 118 ELGEIRPYDETEATLRECL 136
E G IRPY + T + L
Sbjct 3044 EDGVIRPYRLRDGTGNQML 3062
> Hs8923139
Length=442
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 51/133 (38%), Positives = 73/133 (54%), Gaps = 10/133 (7%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEGVWS-LTNIVTKPIEGAQREGV 59
L+ T D EY +R+ E R D GK GV +T I+TKP+EGA++EG
Sbjct 178 LAAITMDKEYQQKRREELSRQPRDFGDSLARGGKGFLRGVVGGVTGIITKPVEGAKKEGA 237
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIK--AEVSKPIGASKFRNERRRKPRMLG 117
GFFKGIGKG+VG++ +P + S +GI+ AE ++ + + R PR++
Sbjct 238 AGFFKGIGKGLVGAVARPTGGIVDMASSTFQGIQRAAESTEEVSS-------LRPPRLIH 290
Query 118 ELGEIRPYDETEA 130
E G IRPYD E+
Sbjct 291 EDGIIRPYDRQES 303
> At1g48090
Length=4099
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 46/134 (34%), Positives = 72/134 (53%), Gaps = 5/134 (3%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEGVW-SLTNIVTKPIEGAQREGV 59
++ + D ++I RQR+ + D G + +G++ +T I+TKP+EGA+ GV
Sbjct 3788 IAALSMDKKFIQSRQRQENKGVEDFGDIIREGGGALAKGLFRGVTGILTKPLEGAKSSGV 3847
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNE---RRRKPRML 116
GF G GKGI+G+ +P+ V +S T G A K I A+ +E RRR PR +
Sbjct 3848 EGFVSGFGKGIIGAAAQPVSGVLDLLSKTTEGANAMRMK-IAAAITSDEQLLRRRLPRAV 3906
Query 117 GELGEIRPYDETEA 130
G +RPY++ A
Sbjct 3907 GADSLLRPYNDYRA 3920
> 7304192
Length=3242
Score = 67.4 bits (163), Expect = 9e-12, Method: Composition-based stats.
Identities = 45/132 (34%), Positives = 66/132 (50%), Gaps = 6/132 (4%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEG-VWSLTNIVTKPIEGAQREGV 59
L+ TFD +Y +R++ + +G + K + G V +T +VTKP+ GA+ GV
Sbjct 2984 LAALTFDEDYQKKRRQGIQNKPKNFHEGLARSSKGLVMGFVDGVTGVVTKPVTGARDNGV 3043
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNERRRKPRMLGEL 119
GFFKG+GKG +G + +P V D G V + AS+ +R R PR
Sbjct 3044 EGFFKGLGKGAIGLVARPT----AGVVDFASGSFEAVKRAADASE-DVKRMRPPRFQHYD 3098
Query 120 GEIRPYDETEAT 131
+RPY EAT
Sbjct 3099 FVLRPYCLMEAT 3110
> At4g17120
Length=1661
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 36/114 (31%), Positives = 58/114 (50%), Gaps = 3/114 (2%)
Query 20 RNTASMRDGFLSAGKNIGEGV-WSLTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPL 78
R + D + + + +GV + ++ +VTKP+E A+ G+ GF G+G+ +G +V+P+
Sbjct 1536 RRITGVGDAIVQGTEALAQGVAFGVSGVVTKPVESARENGILGFAHGVGRAFLGFIVQPV 1595
Query 79 DKVGQAVSDVTRGIKAEVSKPIGASKFRN--ERRRKPRMLGELGEIRPYDETEA 130
S GI A S+ + R ER R PR + G +R YDE EA
Sbjct 1596 SGALDFFSLTVDGIGASCSRCLEVLSNRTALERIRNPRAVHADGILREYDEKEA 1649
> Hs22041525
Length=1231
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 70/131 (53%), Gaps = 8/131 (6%)
Query 12 NRRQRER--VRNTASMRDGFLSAGKN-IGEGVWS-LTNIVTKPIEGAQREG-VGGFFKGI 66
NR Q ER +R A+ L AG + + G+ LT+++T +EG + EG V GF G+
Sbjct 969 NRHQSEREYIRYHAATSGEHLVAGIHGLAHGIIGGLTSVITSTVEGVKTEGGVSGFISGL 1028
Query 67 GKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNERRRKPRML-GELGEIRPY 125
GKG+VG++ KP+ S+ + ++ + + + + +R RKPR G G + Y
Sbjct 1029 GKGLVGTVTKPVAGALDFASETAQAVRDTAT--LSGPRTQAQRVRKPRCCTGPQGLLPRY 1086
Query 126 DETEATLRECL 136
E++A +E L
Sbjct 1087 SESQAEGQEQL 1097
> YLL040c
Length=3144
Score = 54.3 bits (129), Expect = 7e-08, Method: Composition-based stats.
Identities = 38/122 (31%), Positives = 57/122 (46%), Gaps = 12/122 (9%)
Query 9 EYINRRQRERVRNTASMRDGFLSAGKNIGEGVWSLTNIVTKPIEGAQREGVGGFFKGIGK 68
+ IN+ R + N+A S +G G L+ I P + Q+EG GF KG+GK
Sbjct 2925 QRINKNNRNALANSAQ------SFASTLGSG---LSGIALDPYKAMQKEGAAGFLKGLGK 2975
Query 69 GIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNERRRKPRMLGELGEIRPYDET 128
GIVG K S++++G+K+ + + R R PR + I+PYD
Sbjct 2976 GIVGLPTKTAIGFLDLTSNLSQGVKSTTTV---LDMQKGCRVRLPRYVDHDQIIKPYDLR 3032
Query 129 EA 130
EA
Sbjct 3033 EA 3034
> SPBC31F10.18c
Length=600
Score = 47.4 bits (111), Expect = 9e-06, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 55/111 (49%), Gaps = 9/111 (8%)
Query 28 GFLSAGKNIGEGVWS-LTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVS 86
G + ++ GV S + + +PI GA+R G+ G KG+GKG+VG KPL + S
Sbjct 386 GVTAGAASLYTGVRSGVRGLALQPIIGARRNGLPGLVKGLGKGLVGFTTKPLVGLFDFAS 445
Query 87 DVTRGIKAEVSKPIGASKFRNERRRKPRMLGELGEIRPYDETEATLRECLG 137
++ G + + + E+ R R++ + G + P+ LRE LG
Sbjct 446 SISEGARNTTTV---FDERHIEKLRLSRLMSDDGVVYPFQ-----LREALG 488
> ECU04g0690
Length=2371
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 46/85 (54%), Gaps = 11/85 (12%)
Query 43 LTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGA 102
+ I T PIEGA +GV G KG+GKGI+G+ +P+ +V V+ ++ IK +
Sbjct 2200 IAGIATSPIEGAS-QGVTGVVKGLGKGILGAFTRPIVEVADLVTGISDTIKVSMDG---- 2254
Query 103 SKFRNERRRKPRMLGELGEIRPYDE 127
R +R + PR G +G YDE
Sbjct 2255 ---RIKRLQYPRPRGFVGW---YDE 2273
Score = 27.7 bits (60), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 58 GVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGI 92
G G F I +GI G P++ Q V+ V +G+
Sbjct 2188 GTGDLFDSITRGIAGIATSPIEGASQGVTGVVKGL 2222
> CE13443
Length=3212
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 43/131 (32%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIG-EGVWSLTNIVTKPIEGAQREGV 59
++ TFD +Y+ +RQ + R S +G K +G V +T +VTKPIEGA++EG
Sbjct 2971 VAALTFDDDYMKKRQEDLNRKPQSFGEGMARGLKGLGMGVVGGITGVVTKPIEGAKQEGG 3030
Query 60 GGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNERRRKPRMLGEL 119
GF KG+GKG++G + +P+ V S ++A + + R PR+L E
Sbjct 3031 FGFVKGVGKGLIGVVTRPVSGVVDFASGTMNSVRA-----VAGTNREAGPLRPPRVLRED 3085
Query 120 GEIRPYDETEA 130
++PY +A
Sbjct 3086 KIVKPYSSGDA 3096
> 7294545
Length=1902
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 43 LTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGI 92
+T+IV +GA +GV GF G+GKG+VG++ KP+ V S+ +
Sbjct 1788 VTSIVRHTYDGATSDGVPGFLSGLGKGLVGTVTKPIIGVLDLASETASAV 1837
> Hs22048785
Length=1687
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 45/102 (44%), Gaps = 10/102 (9%)
Query 1 LSTFTFDSEYINRRQRERVRNTASMRDGFLSAGKNIGEGVW-SLTNIVTKPIEGAQREG- 58
+ + D E+ NR++ R + S+ +G +G + ++ IV +P++ Q+
Sbjct 1388 MDRLSLDEEHYNRQEEWRRQLPESLGEGLRQGLSRLGISLLGAIAGIVDQPMQNFQKTSE 1447
Query 59 --------VGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGI 92
G G+GKGI+G KP+ + VS GI
Sbjct 1448 AQASAGHKAKGVISGVGKGIMGVFTKPIGGAAELVSQTGYGI 1489
> SPBC16C6.02c
Length=3131
Score = 36.6 bits (83), Expect = 0.016, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 39/93 (41%), Gaps = 6/93 (6%)
Query 47 VTKPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFR 106
+ KP + G F KG GKG++G KP + S+V+ GI+
Sbjct 2812 LKKPFTDPKNNSAGKFLKGFGKGMLGLATKPAIGLLDMTSNVSEGIRNSTDVRTNPEI-- 2869
Query 107 NERRRKPRML---GELGEIRPYDETEATLRECL 136
++ R PR + G + +PY+ + CL
Sbjct 2870 -DKVRVPRYVEFGGLIVPFKPYESLGKYMLSCL 2901
> At4g30160
Length=974
Score = 30.8 bits (68), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 6/47 (12%)
Query 22 TASMRD---GFLSAGKNIGEGVWSLTNIVTKPIEGAQREGVGGFFKG 65
+ SMRD F AG+ G +W + N + PI + +G FF G
Sbjct 2 SVSMRDLDPAFQGAGQKAGIEIWRIENFIPTPIP---KSSIGKFFTG 45
> At2g36780
Length=496
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 41/88 (46%), Gaps = 7/88 (7%)
Query 38 EGVWSLTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPL-----DKVGQAVSDVTRGI 92
EG+ S ++T P+ G Q + + G+ + + + DK+G V G+
Sbjct 382 EGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWGEEDKIGVLVD--KEGV 439
Query 93 KAEVSKPIGASKFRNERRRKPRMLGELG 120
K V + +G S ERRR+ + LGEL
Sbjct 440 KKAVEELMGDSDDAKERRRRVKELGELA 467
> At2g36770
Length=496
Score = 28.5 bits (62), Expect = 4.0, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 41/86 (47%), Gaps = 7/86 (8%)
Query 38 EGVWSLTNIVTKPIEGAQREGVGGFFKGIGKGIVGSLVKPL-----DKVGQAVSDVTRGI 92
EG+ S ++T P+ G Q + + G+ + + + +K+G V G+
Sbjct 382 EGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEEVMKWGEEEKIGVLVD--KEGV 439
Query 93 KAEVSKPIGASKFRNERRRKPRMLGE 118
K V + +GAS ERRR+ + LGE
Sbjct 440 KKAVEELMGASDDAKERRRRVKELGE 465
> Hs4503297
Length=1279
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Query 9 EYINRRQRERVRNTASMRDGFLSAGKNIGEGVWSLTNIVTKPIEGAQREGVGGFFK 64
+Y +R++ + V+ T + L+AG + G W+L N + I+ Q+E + G +K
Sbjct 147 DYYSRKEEQEVQATLESEEVDLNAGLH---GNWTLENAKARLIQYFQKEKIQGEYK 199
> At2g36760
Length=496
Score = 28.1 bits (61), Expect = 5.4, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 91 GIKAEVSKPIGASKFRNERRRKPRMLGELG 120
G+K V + +G S ERR++ R LGEL
Sbjct 438 GVKKAVDEIMGESDEAKERRKRVRELGELA 467
> At2g36750
Length=491
Score = 27.7 bits (60), Expect = 7.0, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 42/88 (47%), Gaps = 7/88 (7%)
Query 38 EGVWSLTNIVTKPIEGAQ----REGVGGFFKGIGKGIVGSLV-KPLDKVGQAVSDVTRGI 92
EG+ S ++T P+ G Q + V G+ G+ S+ +K+G V G+
Sbjct 377 EGITSGVPLLTWPLFGDQFCNEKLAVQILKAGVRAGVEESMRWGEEEKIGVLVD--KEGV 434
Query 93 KAEVSKPIGASKFRNERRRKPRMLGELG 120
K V + +G S ERR++ + LGEL
Sbjct 435 KKAVEELMGDSNDAKERRKRVKELGELA 462
> 7301939
Length=638
Score = 27.7 bits (60), Expect = 7.1, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 37/91 (40%), Gaps = 2/91 (2%)
Query 49 KPIEGAQREGVGGFFKGIGKGIVGSLVKPLDKVGQAVSDVTRGIKAEVSKPIGASKFRNE 108
K I G G+G F G G + S+ L K QA S V+ I E+S+
Sbjct 2 KLIRGTTLLGIGILFVGCGAFSLPSVYDVLSKDQQATSFVSASIAEEISREYLKYHRTGI 61
Query 109 RRRKPRMLGELGEIRPYDETEATLRECLGLL 139
+ R LG+ +IR +A E + L
Sbjct 62 ETERLRQLGK--DIRSSHSKKAIFTESMADL 90
> At3g19190
Length=1814
Score = 27.3 bits (59), Expect = 9.1, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 43/84 (51%), Gaps = 4/84 (4%)
Query 14 RQRERVRNTA--SMRDGFLSAGKNIGEGVW-SLTNIVTKPIEGAQR-EGVGGFFKGIGKG 69
R + VR+ + + G L A ++IG+G+ + + +V P++ QR +G G F + +G
Sbjct 1685 RTKTNVRHNQPRNAKQGMLKACESIGDGIGKTASALVRTPLKKYQRGDGAGSAFATVVQG 1744
Query 70 IVGSLVKPLDKVGQAVSDVTRGIK 93
+ + + P +AV GI+
Sbjct 1745 VPTAAIAPASACARAVHSALVGIR 1768
Lambda K H
0.317 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40