bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5845_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
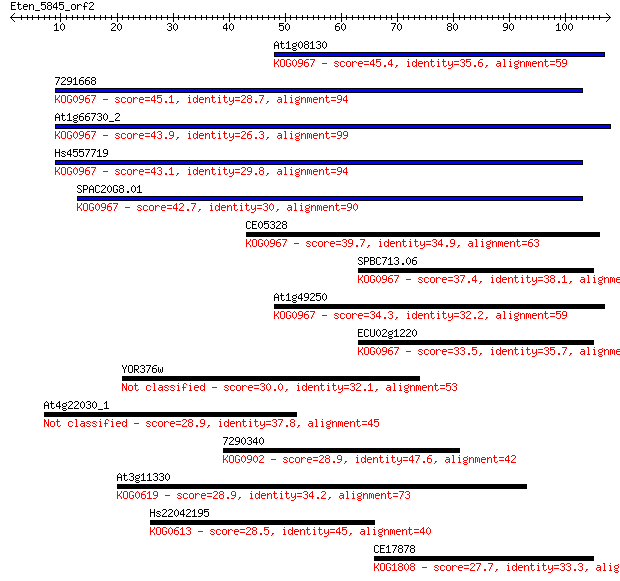
At1g08130 45.4 2e-05
7291668 45.1 3e-05
At1g66730_2 43.9 8e-05
Hs4557719 43.1 1e-04
SPAC20G8.01 42.7 1e-04
CE05328 39.7 0.001
SPBC713.06 37.4 0.007
At1g49250 34.3 0.051
ECU02g1220 33.5 0.085
YOR376w 30.0 1.1
At4g22030_1 28.9 2.3
7290340 28.9 2.3
At3g11330 28.9 2.5
Hs22042195 28.5 3.4
CE17878 27.7 6.0
> At1g08130
Length=794
Score = 45.4 bits (106), Expect = 2e-05, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 48 TLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVSDN 106
T IL N R ++ P+ L A VY N AP E G+G+S + KA++E +G +++
Sbjct 197 TDILCNMLRTVIATTPEDLVATVYLSANEIAPAHEGVELGIGESTIIKAISEAFGRTED 255
> 7291668
Length=692
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 44/94 (46%), Gaps = 12/94 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
K ++ L L R+F V+ E GR+ K L+N F ++ P+ L
Sbjct 65 KDKKVTPYLALARTFQVIEETKGRL------------KMIDTLSNFFCSVMLVSPEDLVP 112
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
+VY +N+ AP + E GV ++ L KA+ + G
Sbjct 113 SVYLSINQLAPAYEGLELGVAETTLMKAICKATG 146
> At1g66730_2
Length=684
Score = 43.9 bits (102), Expect = 8e-05, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 12/99 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
++ Q + L R+F+ + G+I+ + +L N FR L P+ +
Sbjct 27 REGQPAPYIHLVRTFASVESEKGKIKAMS------------MLCNMFRSLFALSPEDVLP 74
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVSDNS 107
AVY C N+ A D + E +G S++ A+ E G+S ++
Sbjct 75 AVYLCTNKIAADHENIELNIGGSLISSALEEACGISRST 113
> Hs4557719
Length=919
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 40/94 (42%), Gaps = 12/94 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
K Q+ L + R+F + E R+ ++ L+N R +V P L
Sbjct 282 KPGQKVPYLAVARTFEKIEEVSARLRMVET------------LSNLLRSVVALSPPDLLP 329
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
+Y LN P E GVGD +L KAVA+ G
Sbjct 330 VLYLSLNHLGPPQQGLELGVGDGVLLKAVAQATG 363
> SPAC20G8.01
Length=474
Score = 42.7 bits (99), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 47/92 (51%), Gaps = 9/92 (9%)
Query 13 RGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLI--LANAFRILVFYCPQALAAAV 70
R +L +F+ + +A +IE +SK+ +I + F ++ P L A V
Sbjct 139 RNDKLKGHATFAEMVKAFTKIEN-------TSKRLEIIDIMGTYFFGILRDHPSDLLACV 191
Query 71 YFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
Y +N+ PD+ E G+G+SI+ KA+ E+ G
Sbjct 192 YLSINKLGPDYSGLELGIGESIIMKAIGESTG 223
> CE05328
Length=847
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 43 SSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
S KK+ LA F ++ + P L A VY +N+ P + E GV ++ L KAVA+ G
Sbjct 109 SGKKKVDELAQFFTKVLDFSPDDLTACVYMSVNQLGPSYEGLELGVAENSLIKAVAKATG 168
Query 103 VSD 105
++
Sbjct 169 RTE 171
> SPBC713.06
Length=774
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 63 PQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVS 104
P +L A V+ C N AP+F GVG ++ KA+ E G++
Sbjct 197 PDSLIATVWLCTNSIAPNFYGKNLGVGPAMYSKALKEVCGIT 238
> At1g49250
Length=657
Score = 34.3 bits (77), Expect = 0.051, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 48 TLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGD-SILFKAVAETYGVSDN 106
T IL N R ++ P L VY N AP + G+G S + KA++E +G +++
Sbjct 65 THILCNMLRTVIATTPDDLLPTVYLAANEIAPAHEGIKLGMGKGSYIIKAISEAFGRTES 124
> ECU02g1220
Length=589
Score = 33.5 bits (75), Expect = 0.085, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 63 PQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVS 104
P ++A A+Y + P++ + E GVG++ L VAE G S
Sbjct 41 PPSMAPALYLSIATVYPEYFNKELGVGENALVGIVAEVTGKS 82
> YOR376w
Length=122
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 21 RSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQAL-----AAAVYFC 73
R+FSVL + + K +G+ + +L++ + +FYCP AL A +FC
Sbjct 35 RNFSVLIFIM--VPDEKENGAAADNSFSLLIGRGVVLFLFYCPTALKMHGPVPAHWFC 90
> At4g22030_1
Length=386
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Query 7 RKKQRQRG---QRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLIL 51
R++Q++RG Q L R +VL E + R+E KN G + +L+L
Sbjct 70 RQRQKKRGCDDQVLQRSRLMAVLEEVIDRVEMHKNIGEQRNNWNSLLL 117
> 7290340
Length=2113
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 21/42 (50%), Gaps = 10/42 (23%)
Query 39 SGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPD 80
SG G QTL FR YCPQ AA V FCL+ A D
Sbjct 68 SGGGGKAVQTL-----FR----YCPQENAAGV-FCLDTRAQD 99
> At3g11330
Length=537
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 25/74 (33%), Positives = 36/74 (48%), Gaps = 7/74 (9%)
Query 20 GRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAP 79
GR +L EA GRI+GL ++K + + N + F+ A+AA V+ A
Sbjct 207 GRKLRLLPEAFGRIQGLLVLNLSNNKLERSYMLNIRCGISFWLLPAIAADVH------AS 260
Query 80 DFLD-AEAGVGDSI 92
FLD +E V SI
Sbjct 261 SFLDSSEVYVQQSI 274
> Hs22042195
Length=652
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 26/43 (60%), Gaps = 3/43 (6%)
Query 26 LTEALGRIEGLKNSGSGSSKKQTLI---LANAFRILVFYCPQA 65
LT+ L + GL +SG + +TL +A++FRI V CPQA
Sbjct 402 LTQLLIPVAGLSDSGLYTVVLRTLQGKEVAHSFRIRVAACPQA 444
> CE17878
Length=2030
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 66 LAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVS 104
L + C++ P L E GVG + + +AVA+ GV+
Sbjct 368 LMERIVVCVSHNEPLLLVGETGVGKTSVVQAVADLIGVT 406
Lambda K H
0.322 0.137 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40