bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5803_orf1
Length=118
Score E
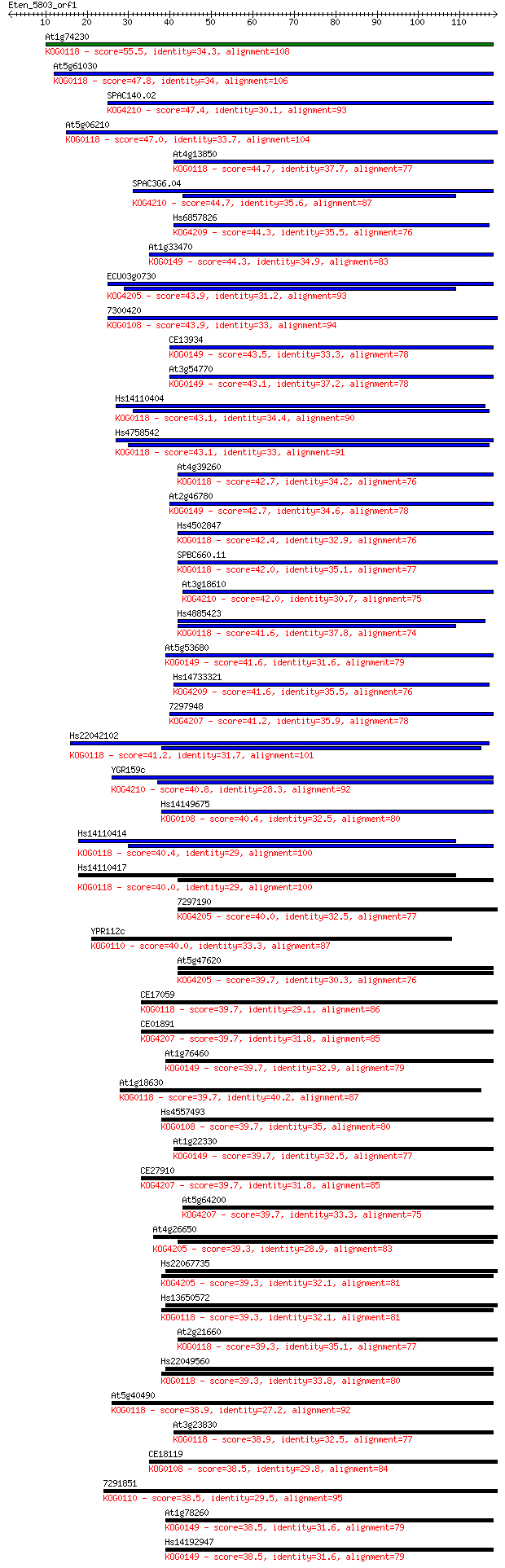
Sequences producing significant alignments: (Bits) Value
At1g74230 55.5 3e-08
At5g61030 47.8 5e-06
SPAC140.02 47.4 6e-06
At5g06210 47.0 9e-06
At4g13850 44.7 4e-05
SPAC3G6.04 44.7 4e-05
Hs6857826 44.3 6e-05
At1g33470 44.3 6e-05
ECU03g0730 43.9 6e-05
7300420 43.9 7e-05
CE13934 43.5 9e-05
At3g54770 43.1 1e-04
Hs14110404 43.1 1e-04
Hs4758542 43.1 1e-04
At4g39260 42.7 1e-04
At2g46780 42.7 2e-04
Hs4502847 42.4 2e-04
SPBC660.11 42.0 2e-04
At3g18610 42.0 3e-04
Hs4885423 41.6 3e-04
At5g53680 41.6 4e-04
Hs14733321 41.6 4e-04
7297948 41.2 5e-04
Hs22042102 41.2 5e-04
YGR159c 40.8 7e-04
Hs14149675 40.4 8e-04
Hs14110414 40.4 8e-04
Hs14110417 40.0 0.001
7297190 40.0 0.001
YPR112c 40.0 0.001
At5g47620 39.7 0.001
CE17059 39.7 0.001
CE01891 39.7 0.001
At1g76460 39.7 0.001
At1g18630 39.7 0.001
Hs4557493 39.7 0.001
At1g22330 39.7 0.001
CE27910 39.7 0.001
At5g64200 39.7 0.002
At4g26650 39.3 0.002
Hs22067735 39.3 0.002
Hs13650572 39.3 0.002
At2g21660 39.3 0.002
Hs22049560 39.3 0.002
At5g40490 38.9 0.002
At3g23830 38.9 0.003
CE18119 38.5 0.003
7291851 38.5 0.003
At1g78260 38.5 0.003
Hs14192947 38.5 0.003
> At1g74230
Length=289
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/108 (34%), Positives = 56/108 (51%), Gaps = 8/108 (7%)
Query 10 YLQRRGRELNREINLLNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKS 69
+L + GR ++ + + S + S +K+FVGG++YST + L E F++ G +
Sbjct 3 FLSKVGRLFSQTSSHVTASSSMLQSIRCMSSSKIFVGGISYSTDEFGLREAFSKYGEV-- 60
Query 70 IIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
VD I D E S RG AFV F E A A+Q DG+ ++GR
Sbjct 61 -----VDAKIIVDRETGRS-RGFAFVTFTSTEEASNAMQLDGQDLHGR 102
> At5g61030
Length=309
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 56/107 (52%), Gaps = 11/107 (10%)
Query 12 QRRGRELNREINLLNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSII 71
Q ++LN +++L + + +A + S +K+F+GG+AYS L E F + G +
Sbjct 13 QTTNKQLNAQVSLSSPSLFQA--IRCMSSSKLFIGGMAYSMDEDSLREAFTKYGEV---- 66
Query 72 IPQVDRSIPPDYEGAYSVRGIAFVEFGDAETAKKALQC-DGRLVNGR 117
VD + D E S RG FV F +E A A+Q DGR ++GR
Sbjct 67 ---VDTRVILDRETGRS-RGFGFVTFTSSEAASSAIQALDGRDLHGR 109
> SPAC140.02
Length=500
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 47/93 (50%), Gaps = 15/93 (16%)
Query 25 LNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYE 84
DQ+SE + VFVG L+++ + DL F G+I+SI +P +S
Sbjct 357 FGDQLSEPSDT-------VFVGNLSFNATEDDLSTAFGGCGDIQSIRLPTDPQS------ 403
Query 85 GAYSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
++G +V F D ++AKK ++ +G + GR
Sbjct 404 --GRLKGFGYVTFSDIDSAKKCVEMNGHFIAGR 434
> At5g06210
Length=146
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 57/108 (52%), Gaps = 16/108 (14%)
Query 15 GRELNREINLLNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNI--KSIII 72
GR L + + L+N S +K+F+GGL++ T+ L E F++ G + I++
Sbjct 9 GRHL-KSVCLINSSASCFFTQRRGVASKLFIGGLSFCTTEQGLSEAFSKCGQVVEAQIVM 67
Query 73 PQV-DRSIPPDYEGAYSVRGIAFVEFGDAETAKKAL-QCDGRLVNGRT 118
+V DRS +G FV F A+ A+KAL + +G+ +NGRT
Sbjct 68 DRVSDRS-----------KGFGFVTFASADEAQKALMEFNGQQLNGRT 104
> At4g13850
Length=158
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 41/78 (52%), Gaps = 9/78 (11%)
Query 41 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDA 100
TK+F+GGL++ T L + FA G++ VD + D E S RG FV F D
Sbjct 35 TKLFIGGLSWGTDDASLRDAFAHFGDV-------VDAKVIVDRETGRS-RGFGFVNFNDE 86
Query 101 ETAKKAL-QCDGRLVNGR 117
A A+ + DG+ +NGR
Sbjct 87 GAATAAISEMDGKELNGR 104
> SPAC3G6.04
Length=369
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 49/98 (50%), Gaps = 11/98 (11%)
Query 31 EAAGMSTKSLTKVFVGGLAYSTSRYDLCELFA--------EVG--NIKSIIIPQVDR-SI 79
E+ S +S ++VG L++ T++ L + F EV NIK I Q+ R +
Sbjct 95 ESQEESKRSPWGIWVGNLSFHTTKEILTDFFVRETSEMIKEVSEENIKPITTEQITRIHM 154
Query 80 PPDYEGAYSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
P E + +G A+V+F + K ALQC + +NGR
Sbjct 155 PMSKEKRFQNKGFAYVDFATEDALKLALQCSEKALNGR 192
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 8/66 (12%)
Query 43 VFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAET 102
+FVG L + T+ DL E F +VG I+ + + +E +G FV+F D +T
Sbjct 230 LFVGNLDFETTDADLKEHFGQVGQIRRVRLMT--------FEDTGKCKGFGFVDFPDIDT 281
Query 103 AKKALQ 108
KA++
Sbjct 282 CMKAME 287
> Hs6857826
Length=305
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 45/77 (58%), Gaps = 8/77 (10%)
Query 41 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDA 100
TKV +G L + ++ + E+F+ G IK I +P V+R P + +G A+VEF +
Sbjct 161 TKVHIGRLTRNVTKDHIMEIFSTYGKIKMIDMP-VERMHP------HLSKGYAYVEFENP 213
Query 101 ETAKKALQ-CDGRLVNG 116
+ A+KAL+ DG ++G
Sbjct 214 DEAEKALKHMDGGQIDG 230
> At1g33470
Length=117
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 10/84 (11%)
Query 35 MSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQVDRSIPPDYEGAYSVRGIA 93
M + TKVFVGGLA+ T + L F + G+I ++++I D+S + +G
Sbjct 1 MEDTTFTKVFVGGLAWETHKVSLRNYFEQFGDIVEAVVI--TDKS-------SGRSKGYG 51
Query 94 FVEFGDAETAKKALQCDGRLVNGR 117
FV F D E A+KA +++GR
Sbjct 52 FVTFCDPEAAQKACVDPAPVIDGR 75
> ECU03g0730
Length=293
Score = 43.9 bits (102), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 50/97 (51%), Gaps = 13/97 (13%)
Query 25 LNDQVSEAAGMSTK----SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIP 80
+ ++ S +G ++ SLT VF+ G++Y + YDL ++G + + IP +
Sbjct 68 IEEETSSGSGEESRRENPSLT-VFIKGISYDLTEYDLKSEMEKIGKVARVGIPMTN---- 122
Query 81 PDYEGAYSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
D++ +G +VEF E KKAL+ DG + GR
Sbjct 123 -DHK---RNKGFGYVEFCKEEDVKKALKLDGTVFLGR 155
Score = 30.8 bits (68), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query 29 VSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYS 88
V+ A + K ++V + Y + DL + F +G + V S+P D +
Sbjct 159 VNMAHPRANKERHTIYVSNIPYECDKRDLKKYFEGMGEV-------VGMSLPYDRDNN-R 210
Query 89 VRGIAFVEFGDAETAKKALQ 108
++G FV+FG+ E ++ L+
Sbjct 211 LKGYGFVDFGNKEDYERVLK 230
> 7300420
Length=399
Score = 43.9 bits (102), Expect = 7e-05, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 48/95 (50%), Gaps = 10/95 (10%)
Query 25 LNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYE 84
+ D+ E + M KS+ VFVG + Y + L E+F+EVG + S+ + S P
Sbjct 1 MADKAQEQSIMD-KSMRSVFVGNIPYEATEEKLKEIFSEVGPVLSLKLVFDRESGKP--- 56
Query 85 GAYSVRGIAFVEFGDAETAKKALQ-CDGRLVNGRT 118
+G F E+ D ETA A++ +G + GRT
Sbjct 57 -----KGFGFCEYKDQETALSAMRNLNGYEIGGRT 86
> CE13934
Length=248
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 8/78 (10%)
Query 40 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGD 99
TK+FVGGL Y TS L E F + G+I+ ++ DR+ RG FV D
Sbjct 34 FTKIFVGGLPYHTSDKTLHEYFEQFGDIEEAVV-ITDRNT-------QKSRGYGFVTMKD 85
Query 100 AETAKKALQCDGRLVNGR 117
+A++A + +++GR
Sbjct 86 RASAERACKDPNPIIDGR 103
> At3g54770
Length=261
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 14/81 (17%)
Query 40 LTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQ--VDRSIPPDYEGAYSVRGIAFVE 96
LTKVFVGGLA+ T + + + F + G+I +++II RS +G FV
Sbjct 16 LTKVFVGGLAWDTHKEAMYDHFIKYGDILEAVIISDKLTRRS-----------KGYGFVT 64
Query 97 FGDAETAKKALQCDGRLVNGR 117
F DA+ A +A + ++NGR
Sbjct 65 FKDAKAATRACEDSTPIINGR 85
> Hs14110404
Length=331
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 48/97 (49%), Gaps = 21/97 (21%)
Query 27 DQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDY 83
DQ++ A + + K+FVGGL++ TS+ DL + F + G + I P RS
Sbjct 56 DQIN--ASKNEEDAGKMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRS----- 108
Query 84 EGAYSVRGIAFVEFGDAETAKKAL-----QCDGRLVN 115
RG F+ F DA + +K L + DGR+++
Sbjct 109 ------RGFGFILFKDAASVEKVLDQKEHRLDGRVID 139
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 41/87 (47%), Gaps = 9/87 (10%)
Query 31 EAAGMSTKSLTKVFVGGL-AYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSV 89
+A M + K+FVGGL S + + E F E G I++I +P +D +
Sbjct 142 KAMAMKKDPVKKIFVGGLNPESPTEEKIREYFGEFGEIEAIELP-MDPKL-------NKR 193
Query 90 RGIAFVEFGDAETAKKALQCDGRLVNG 116
RG F+ F + E KK L+ V+G
Sbjct 194 RGFVFITFKEEEPVKKVLEKKFHTVSG 220
> Hs4758542
Length=284
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 21/99 (21%)
Query 27 DQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDY 83
DQ++ A + + K+FVGGL++ TS+ DL + F + G + I P RS
Sbjct 56 DQIN--ASKNEEDAGKMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRS----- 108
Query 84 EGAYSVRGIAFVEFGDAETAKKAL-----QCDGRLVNGR 117
RG F+ F DA + +K L + DGR+++ +
Sbjct 109 ------RGFGFILFKDAASVEKVLDQKEHRLDGRVIDPK 141
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 41/88 (46%), Gaps = 9/88 (10%)
Query 30 SEAAGMSTKSLTKVFVGGL-AYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYS 88
+A M + K+FVGGL S + + E F E G I++I +P +D +
Sbjct 141 KKAMAMKKDPVKKIFVGGLNPESPTEEKIREYFGEFGEIEAIELP-MDPKL-------NK 192
Query 89 VRGIAFVEFGDAETAKKALQCDGRLVNG 116
RG F+ F + E KK L+ V+G
Sbjct 193 RRGFVFITFKEEEPVKKVLEKKFHTVSG 220
> At4g39260
Length=169
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 43/77 (55%), Gaps = 9/77 (11%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
+ FVGGLA++T+ DL F++ G++ +D I D E S RG FV F D +
Sbjct 7 RCFVGGLAWATNDEDLQRTFSQFGDV-------IDSKIINDRESGRS-RGFGFVTFKDEK 58
Query 102 TAKKAL-QCDGRLVNGR 117
+ A+ + +G+ ++GR
Sbjct 59 AMRDAIEEMNGKELDGR 75
> At2g46780
Length=304
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 43/79 (54%), Gaps = 10/79 (12%)
Query 40 LTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQVDRSIPPDYEGAYSVRGIAFVEFG 98
LTK+FVGGLA+ T R + F + G I ++++I D++ +G FV F
Sbjct 21 LTKIFVGGLAWETQRDTMRRYFEQFGEIVEAVVI--TDKNTG-------RSKGYGFVTFK 71
Query 99 DAETAKKALQCDGRLVNGR 117
+AE A +A Q +++GR
Sbjct 72 EAEAAMRACQNMNPVIDGR 90
> Hs4502847
Length=172
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 43/77 (55%), Gaps = 9/77 (11%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+FVGGL++ T+ L ++F++ G I +++ + DR RG FV F + +
Sbjct 7 KLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVK-DRETQR-------SRGFGFVTFENID 58
Query 102 TAKKALQC-DGRLVNGR 117
AK A+ +G+ V+GR
Sbjct 59 DAKDAMMAMNGKSVDGR 75
> SPBC660.11
Length=348
Score = 42.0 bits (97), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 18/82 (21%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIP-----QVDRSIPPDYEGAYSVRGIAFVE 96
+VFVG L+ ST + ++ LF VG ++ + IP + R +P GIAFV
Sbjct 45 RVFVGRLSTSTKKSEIRSLFETVGTVRKVTIPFRRVRRGTRLVP---------SGIAFVT 95
Query 97 FGDAETAKKALQCDGRLVNGRT 118
F + E KA++ +NG+T
Sbjct 96 FNNQEDVDKAIET----LNGKT 113
> At3g18610
Length=636
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 37/75 (49%), Gaps = 9/75 (12%)
Query 43 VFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAET 102
+F G L+Y +R D+ F E G + + + D S +G +EF E
Sbjct 386 LFAGNLSYQIARSDIENFFKEAGEVVDVRLSSFDDG---------SFKGYGHIEFASPEE 436
Query 103 AKKALQCDGRLVNGR 117
A+KAL+ +G+L+ GR
Sbjct 437 AQKALEMNGKLLLGR 451
> Hs4885423
Length=420
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 28/79 (35%), Positives = 41/79 (51%), Gaps = 13/79 (16%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+F+GGL++ TS+ DL E + G + VD +I D S RG FV F DA
Sbjct 149 KMFIGGLSWDTSKKDLTEYLSRFGEV-------VDCTIKTDPVTGRS-RGFGFVLFKDAA 200
Query 102 TAKKALQ-----CDGRLVN 115
+ K L+ DG+L++
Sbjct 201 SVDKVLELKEHKLDGKLID 219
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 33/67 (49%), Gaps = 8/67 (11%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
KVFVGGL+ TS + E F G I++I +P ++ RG F+ + D E
Sbjct 234 KVFVGGLSPDTSEEQIKEYFGAFGEIENIELPMDTKT--------NERRGFCFITYTDEE 285
Query 102 TAKKALQ 108
KK L+
Sbjct 286 PVKKLLE 292
> At5g53680
Length=169
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 14/82 (17%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSVRGIAFV 95
+ TK++VGGL ++T + L F G I + + + DRS +G FV
Sbjct 11 TFTKIYVGGLPWTTRKEGLINFFKRFGEIIHVNVVCDRETDRS-----------QGYGFV 59
Query 96 EFGDAETAKKALQCDGRLVNGR 117
F DAE+A +A + + GR
Sbjct 60 TFKDAESATRACKDPNPTIEGR 81
> Hs14733321
Length=305
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 44/77 (57%), Gaps = 8/77 (10%)
Query 41 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDA 100
TKV +G L + ++ + E+F+ G IK I +P V+R P + +G A VEF +
Sbjct 161 TKVHIGRLTRNVTKDHIMEIFSTYGKIKMIDMP-VERMHP------HLSKGYACVEFENP 213
Query 101 ETAKKALQ-CDGRLVNG 116
+ A+KAL+ DG ++G
Sbjct 214 DEAEKALKHMDGGQIDG 230
> 7297948
Length=195
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 39/79 (49%), Gaps = 9/79 (11%)
Query 40 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGD 99
+ + V L Y T+ DL +F G + I IP+ DR RG AFV F D
Sbjct 22 MVSLKVDNLTYRTTPEDLRRVFERCGEVGDIYIPR-DRYT-------RESRGFAFVRFYD 73
Query 100 AETAKKALQC-DGRLVNGR 117
A+ AL+ DGR+++GR
Sbjct 74 KRDAEDALEAMDGRMLDGR 92
> Hs22042102
Length=378
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 49/101 (48%), Gaps = 16/101 (15%)
Query 16 RELNREINLLNDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQV 75
R ++RE D V A ++ K K+FVGG+ T Y+L + F + G I++I + +
Sbjct 109 RAVSRE-----DSVKPGAHLTVK---KIFVGGIKEDTEEYNLRDYFEKYGKIETIEVME- 159
Query 76 DRSIPPDYEGAYSVRGIAFVEFGDAETAKKALQCDGRLVNG 116
DR + RG AFV F D +T K + +NG
Sbjct 160 DRQ-------SGKKRGFAFVTFDDHDTVDKIVVQKYHTING 193
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 19/85 (22%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSVRGIAF 94
+ L K+F+GGL++ T+ L E F + G + ++ PQ RS RG F
Sbjct 32 EQLRKLFIGGLSFETTDDSLREHFEKWGTLTDCVVMRDPQTKRS-----------RGFGF 80
Query 95 VEFG-----DAETAKKALQCDGRLV 114
V + DA + + DGR+V
Sbjct 81 VTYSCVEEVDAAMCARPHKVDGRVV 105
> YGR159c
Length=414
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 9/93 (9%)
Query 26 NDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEG 85
ND+ + ++ +F+G L+++ R + ELFA+ G + S+ IP + P
Sbjct 252 NDRAKKFGDTPSEPSDTLFLGNLSFNADRDAIFELFAKHGEVVSVRIPTHPETEQP---- 307
Query 86 AYSVRGIAFVEFGDAETAKKALQC-DGRLVNGR 117
+G +V+F + E AKKAL G ++ R
Sbjct 308 ----KGFGYVQFSNMEDAKKALDALQGEYIDNR 336
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 40/85 (47%), Gaps = 15/85 (17%)
Query 37 TKSLTKVFVGGLAYSTSRYDLCELFAEVGNI---KSIIIPQVDRSIPPDYEGAYSVRGIA 93
T+ +FVG L++S L + F +G + + I DRS RG
Sbjct 164 TEEPATIFVGRLSWSIDDEWLKKEFEHIGGVIGARVIYERGTDRS-----------RGYG 212
Query 94 FVEFGDAETAKKALQ-CDGRLVNGR 117
+V+F + A+KA+Q G+ ++GR
Sbjct 213 YVDFENKSYAEKAIQEMQGKEIDGR 237
> Hs14149675
Length=616
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 9/81 (11%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEF 97
+SL VFVG + Y + L ++F+EVG++ S + + P +G F E+
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGSVVSFRLVYDRETGKP--------KGYGFCEY 64
Query 98 GDAETAKKALQ-CDGRLVNGR 117
D ETA A++ +GR +GR
Sbjct 65 QDQETALSAMRNLNGREFSGR 85
> Hs14110414
Length=306
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 47/95 (49%), Gaps = 12/95 (12%)
Query 18 LNREINLLNDQV---SEAAGMSTKS-LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIP 73
++++ + LN +V A M TK + K+FVGGL+ T + E F G ++SI +P
Sbjct 155 MDQKEHKLNGKVIDPKRAKAMKTKEPVKKIFVGGLSPDTPEEKIREYFGGFGEVESIELP 214
Query 74 QVDRSIPPDYEGAYSVRGIAFVEFGDAETAKKALQ 108
+++ RG F+ F + E KK ++
Sbjct 215 MDNKT--------NKRRGFCFITFKEEEPVKKIME 241
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 45/91 (49%), Gaps = 15/91 (16%)
Query 30 SEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGA 86
SEAA + + K+F+GGL++ T++ DL + F++ G + + P RS
Sbjct 87 SEAA-TAQREEWKMFIGGLSWDTTKKDLKDYFSKFGEVVDCTLKLDPITGRS-------- 137
Query 87 YSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
RG FV F ++E+ K + +NG+
Sbjct 138 ---RGFGFVLFKESESVDKVMDQKEHKLNGK 165
> Hs14110417
Length=336
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 47/95 (49%), Gaps = 12/95 (12%)
Query 18 LNREINLLNDQV---SEAAGMSTKS-LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIP 73
++++ + LN +V A M TK + K+FVGGL+ T + E F G ++SI +P
Sbjct 136 MDQKEHKLNGKVIDPKRAKAMKTKEPVKKIFVGGLSPDTPEEKIREYFGGFGEVESIELP 195
Query 74 QVDRSIPPDYEGAYSVRGIAFVEFGDAETAKKALQ 108
+++ RG F+ F + E KK ++
Sbjct 196 MDNKT--------NKRRGFCFITFKEEEPVKKIME 222
Score = 38.1 bits (87), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 39/79 (49%), Gaps = 14/79 (17%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSVRGIAFVEFG 98
K+F+GGL++ T++ DL + F++ G + + P RS RG FV F
Sbjct 79 KMFIGGLSWDTTKKDLKDYFSKFGEVVDCTLKLDPITGRS-----------RGFGFVLFK 127
Query 99 DAETAKKALQCDGRLVNGR 117
++E+ K + +NG+
Sbjct 128 ESESVDKVMDQKEHKLNGK 146
> 7297190
Length=421
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 37/77 (48%), Gaps = 8/77 (10%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+FVGGL++ T++ +L F G+I +D + + E S RG FV F D
Sbjct 8 KLFVGGLSWETTQENLSRYFCRFGDI-------IDCVVMKNNESGRS-RGFGFVTFADPT 59
Query 102 TAKKALQCDGRLVNGRT 118
LQ ++GRT
Sbjct 60 NVNHVLQNGPHTLDGRT 76
> YPR112c
Length=887
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 46/91 (50%), Gaps = 14/91 (15%)
Query 21 EINLLNDQVSEAAGMSTKS---LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIP-QVD 76
++ L + Q S++ TKS K+ V L + +R D+ ELF G +KS+ +P + D
Sbjct 740 QLKLSHRQASQSGNTKTKSNKKSGKIIVKNLPFEATRKDVFELFNSFGQLKSVRVPKKFD 799
Query 77 RSIPPDYEGAYSVRGIAFVEFGDAETAKKAL 107
+ S RG AFVEF + A+ A+
Sbjct 800 K----------SARGFAFVEFLLPKEAENAM 820
> At5g47620
Length=404
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 8/76 (10%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+FVGGLA S + + + FA+ G I +++ R+ P RG F+ + E
Sbjct 107 KIFVGGLASSVTEAEFKKYFAQFGMITDVVVMYDHRTQRP--------RGFGFISYDSEE 158
Query 102 TAKKALQCDGRLVNGR 117
K LQ +NG+
Sbjct 159 AVDKVLQKTFHELNGK 174
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 12/80 (15%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+F+GG+++ TS L + F G + +I + DR+ RG FV F D
Sbjct 7 KLFIGGISWETSEDRLRDYFHSFGEVLEAVIMK-DRATG-------RARGFGFVVFADPN 58
Query 102 TAKKALQ----CDGRLVNGR 117
A++ + DG++V +
Sbjct 59 VAERVVLLKHIIDGKIVEAK 78
> CE17059
Length=346
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 14/89 (15%)
Query 33 AGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSV 89
A + ++L K+FVGGL +T+ + E +++ G I II+ P RS
Sbjct 15 ASLEPENLRKIFVGGLTSNTTDDLMREFYSQFGEITDIIVMRDPTTKRS----------- 63
Query 90 RGIAFVEFGDAETAKKALQCDGRLVNGRT 118
RG FV F A++ +++G+T
Sbjct 64 RGFGFVTFSGKTEVDAAMKQRPHIIDGKT 92
> CE01891
Length=196
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 9/86 (10%)
Query 33 AGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGI 92
A LT + + L+Y T+ DL F G+I + IP+ S +G
Sbjct 11 AAPDINGLTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYS--------RQSKGF 62
Query 93 AFVEFGDAETAKKAL-QCDGRLVNGR 117
FV F + A+ AL + DG+LV+GR
Sbjct 63 GFVRFYERRDAEHALDRTDGKLVDGR 88
> At1g76460
Length=279
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 42/80 (52%), Gaps = 10/80 (12%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQVDRSIPPDYEGAYSVRGIAFVEF 97
+ TKVFVGGLA+ T L + F + G I ++++I D++ +G FV F
Sbjct 22 TFTKVFVGGLAWETQSETLRQHFEQYGEILEAVVI--ADKN-------TGRSKGYGFVTF 72
Query 98 GDAETAKKALQCDGRLVNGR 117
D E A++A +++GR
Sbjct 73 RDPEAARRACADPTPIIDGR 92
> At1g18630
Length=155
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 42/95 (44%), Gaps = 16/95 (16%)
Query 28 QVSEAAGMSTKSLT--KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEG 85
Q S A SLT K+FVGGL+ ST L E F G I VD + D E
Sbjct 21 QASNAMLQGNLSLTPSKIFVGGLSPSTDVELLKEAFGSFGKI-------VDAVVVLDRES 73
Query 86 AYSVRGIAFVEFGDAETAKKALQC------DGRLV 114
S RG FV + E A A+Q DGR++
Sbjct 74 GLS-RGFGFVTYDSIEVANNAMQAMQNKELDGRII 107
> Hs4557493
Length=577
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 9/81 (11%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEF 97
+SL VFVG + Y + L ++F+EVG P V + D E +G F E+
Sbjct 13 RSLRSVFVGNIPYEATEEQLKDIFSEVG-------PVVSFRLVYDRETG-KPKGYGFCEY 64
Query 98 GDAETAKKALQ-CDGRLVNGR 117
D ETA A++ +GR +GR
Sbjct 65 QDQETALSAMRNLNGREFSGR 85
> At1g22330
Length=291
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 43/78 (55%), Gaps = 10/78 (12%)
Query 41 TKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGD 99
TKVFVGGLA+ T ++ F + G I +++II D++ +G FV F D
Sbjct 17 TKVFVGGLAWETPTDEMRRYFDQFGEILEAVII--TDKAT-------GKSKGYGFVTFRD 67
Query 100 AETAKKALQCDGRLVNGR 117
+++A +A+ +++GR
Sbjct 68 SDSATRAVADPNPVIDGR 85
> CE27910
Length=126
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 9/86 (10%)
Query 33 AGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGI 92
A LT + + L+Y T+ DL F G+I + IP+ S +G
Sbjct 11 AAPDINGLTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYS--------RQSKGF 62
Query 93 AFVEFGDAETAKKAL-QCDGRLVNGR 117
FV F + A+ AL + DG+LV+GR
Sbjct 63 GFVRFYERRDAEHALDRTDGKLVDGR 88
> At5g64200
Length=303
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 40/76 (52%), Gaps = 9/76 (11%)
Query 43 VFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAET 102
+ V + + T+ DL LFA+ G + + IP+ R+ RG AFV + +
Sbjct 18 LLVLNITFRTTADDLYPLFAKYGKVVDVFIPRDRRTG--------DSRGFAFVRYKYKDE 69
Query 103 AKKALQ-CDGRLVNGR 117
A KA++ DGR+V+GR
Sbjct 70 AHKAVERLDGRVVDGR 85
> At4g26650
Length=524
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 42/83 (50%), Gaps = 9/83 (10%)
Query 36 STKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFV 95
S L K+F+GG+++ T L E F + G++ +I + DR+ RG F+
Sbjct 3 SASDLGKLFIGGISWDTDEERLQEYFGKYGDLVEAVIMR-DRTTG-------RARGFGFI 54
Query 96 EFGDAETAKKALQCDGRLVNGRT 118
F D A++ + D +++GRT
Sbjct 55 VFADPSVAERVIM-DKHIIDGRT 76
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 31/76 (40%), Gaps = 8/76 (10%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
K+FVGGL S + + F + G I +++ + P RG F+ F E
Sbjct 116 KIFVGGLPSSITEAEFKNYFDQFGTIADVVVMYDHNTQRP--------RGFGFITFDSEE 167
Query 102 TAKKALQCDGRLVNGR 117
+ L +NG+
Sbjct 168 SVDMVLHKTFHELNGK 183
> Hs22067735
Length=268
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFG 98
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 99 DAETAKKALQCDGRLVNGRT 118
D ++ K + VNG
Sbjct 155 DHDSVDKIVIQKYHTVNGHN 174
Score = 35.0 bits (79), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 14/83 (16%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSVRGIAF 94
+ L K+F+GGL++ T+ L F + G + ++ P RS RG F
Sbjct 11 EQLRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGF 59
Query 95 VEFGDAETAKKALQCDGRLVNGR 117
V + E A+ V+GR
Sbjct 60 VTYATVEEVDAAMNARPHKVDGR 82
> Hs13650572
Length=321
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 38/80 (47%), Gaps = 8/80 (10%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFG 98
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 99 DAETAKKALQCDGRLVNGRT 118
D ++ K + VNG
Sbjct 155 DHDSVDKTVIQKYHTVNGHN 174
Score = 34.7 bits (78), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 14/83 (16%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIII---PQVDRSIPPDYEGAYSVRGIAF 94
+ L K+F+GGL++ T+ L F + G + ++ P RS RG F
Sbjct 11 EQLRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGF 59
Query 95 VEFGDAETAKKALQCDGRLVNGR 117
V + E A+ V+GR
Sbjct 60 VTYATVEEVDAAMNARPHKVDGR 82
> At2g21660
Length=176
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 9/78 (11%)
Query 42 KVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDAE 101
+ FVGGLA++T L FA+ G++ +D I D E S RG FV F D +
Sbjct 9 RCFVGGLAWATDDRALETAFAQYGDV-------IDSKIINDRETGRS-RGFGFVTFKDEK 60
Query 102 TAKKALQ-CDGRLVNGRT 118
K A++ +G+ ++GR+
Sbjct 61 AMKDAIEGMNGQDLDGRS 78
> Hs22049560
Length=222
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 38/79 (48%), Gaps = 8/79 (10%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFG 98
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F
Sbjct 103 TVKKIFVGGIKEDTKEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 99 DAETAKKALQCDGRLVNGR 117
D ++ K + VNG
Sbjct 155 DHDSVDKIVIQKYHTVNGH 173
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 8/80 (10%)
Query 38 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEF 97
K L K+F+GGL++ T+ L F + G + ++ + P+ + + RG FV +
Sbjct 11 KQLRKLFIGGLSFETTNESLRSHFEQWGTLMDCVVMR-----DPNTKCS---RGFGFVTY 62
Query 98 GDAETAKKALQCDGRLVNGR 117
E A+ V+GR
Sbjct 63 ATVEEVDAAMNARPHKVDGR 82
> At5g40490
Length=423
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 43/92 (46%), Gaps = 9/92 (9%)
Query 26 NDQVSEAAGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEG 85
+D+ +G S K+FVGGLA T+ + + F + G I +I + ++ P
Sbjct 27 DDKSQPHSGGGVDSAGKIFVGGLARETTSAEFLKHFGKYGEITDSVIMKDRKTGQP---- 82
Query 86 AYSVRGIAFVEFGDAETAKKALQCDGRLVNGR 117
RG FV + D+ K +Q D ++ G+
Sbjct 83 ----RGFGFVTYADSSVVDKVIQ-DNHIIIGK 109
> At3g23830
Length=136
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 9/78 (11%)
Query 41 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFGDA 100
+K+FVGGL++ T L + F G + + ++ D E S RG FV F
Sbjct 35 SKLFVGGLSWGTDDSSLKQAFTSFGEV-------TEATVIADRETGRS-RGFGFVSFSCE 86
Query 101 ETAKKAL-QCDGRLVNGR 117
++A A+ + DG+ +NGR
Sbjct 87 DSANNAIKEMDGKELNGR 104
> CE18119
Length=84
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 44/85 (51%), Gaps = 10/85 (11%)
Query 35 MSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAF 94
M+++ + V+VG + Y + ++ FA VG++ ++ I + P RG AF
Sbjct 1 MASQGFS-VYVGNVPYQGTEEEIGNYFAAVGHVNNVRIVYDRETGRP--------RGFAF 51
Query 95 VEFGDAETAKKAL-QCDGRLVNGRT 118
VEF + A++A+ Q +G NGR
Sbjct 52 VEFSEEAGAQRAVEQLNGVAFNGRN 76
> 7291851
Length=250
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 43/96 (44%), Gaps = 13/96 (13%)
Query 24 LLNDQVSEA-AGMSTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPD 82
+ +DQ+S G S + ++V L + T++ DL F+ G +KSI IP+ R
Sbjct 96 ITSDQLSSLLRGASKTNRHVLYVTNLNFETTKDDLELHFSAAGTVKSIRIPKKRRG---- 151
Query 83 YEGAYSVRGIAFVEFGDAETAKKALQCDGRLVNGRT 118
G AFVE D + + A Q + GR
Sbjct 152 --------GFAFVEMADLSSFQNAFQLHNTELQGRN 179
> At1g78260
Length=302
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 45/80 (56%), Gaps = 10/80 (12%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIIIPQVDRSIPPDYEGAYSVRGIAFVEF 97
+ TKVFVGGLA+ T ++ F + G I +++II D++ G +G FV F
Sbjct 15 TYTKVFVGGLAWETPTDEMRRYFEQFGEILEAVII--TDKNT-----GK--SKGYGFVTF 65
Query 98 GDAETAKKALQCDGRLVNGR 117
++++A +A+ +++GR
Sbjct 66 RESDSATRAVADPNPVIDGR 85
> Hs14192947
Length=216
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 39/79 (49%), Gaps = 8/79 (10%)
Query 39 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIIIPQVDRSIPPDYEGAYSVRGIAFVEFG 98
+ TK+FVGGL Y T+ L + F G+I+ ++ DR G RG FV
Sbjct 9 TFTKIFVGGLPYHTTDASLRKYFEGFGDIEEAVV-ITDRQT-----G--KSRGYGFVTMA 60
Query 99 DAETAKKALQCDGRLVNGR 117
D A++A + +++GR
Sbjct 61 DRAAAERACKDPNPIIDGR 79
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40