bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5792_orf4
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
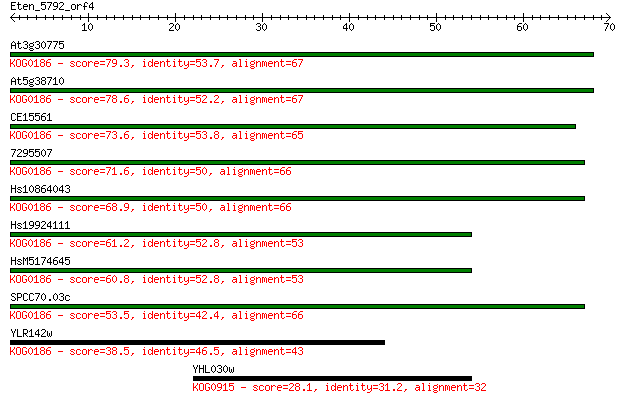
At3g30775 79.3 1e-15
At5g38710 78.6 3e-15
CE15561 73.6 1e-13
7295507 71.6 4e-13
Hs10864043 68.9 2e-12
Hs19924111 61.2 5e-10
HsM5174645 60.8 6e-10
SPCC70.03c 53.5 9e-08
YLR142w 38.5 0.003
YHL030w 28.1 4.0
> At3g30775
Length=499
Score = 79.3 bits (194), Expect = 1e-15, Method: Composition-based stats.
Identities = 36/67 (53%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM DA S GL AGF+V KY+PFGPV IPYLLRRA EN GM+ ++ + + E
Sbjct 430 QLYGMSDALSFGLKRAGFNVSKYMPFGPVATAIPYLLRRAYENRGMMATGAHDRQLMRME 489
Query 61 ITRRVLG 67
+ RR++
Sbjct 490 LKRRLIA 496
> At5g38710
Length=476
Score = 78.6 bits (192), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/67 (52%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM DA S GL AGF+V KY+P+GPV+ IPYL+RRA EN GM+ + + + E
Sbjct 409 QLYGMSDALSFGLKRAGFNVSKYMPYGPVDTAIPYLIRRAYENRGMMSTGALDRQLMRKE 468
Query 61 ITRRVLG 67
+ RRV+
Sbjct 469 LKRRVMA 475
> CE15561
Length=564
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/65 (53%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM D S L AGF VYKY+P+GPVE+ +PYL RRA EN +L A E L E
Sbjct 487 QLYGMCDQVSFSLGQAGFSVYKYLPYGPVEEVLPYLSRRALENGSVLKKANKERDLLWKE 546
Query 61 ITRRV 65
+ RR+
Sbjct 547 LKRRI 551
> 7295507
Length=671
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D + L AG+ YKY+P+GPVE+ +PYL RRA+EN G+L + E R L E
Sbjct 591 QLLGMCDYITFPLGQAGYSAYKYIPYGPVEEVLPYLSRRAQENKGVLKKIKKEKRLLLSE 650
Query 61 ITRRVL 66
I RR++
Sbjct 651 IRRRLM 656
> Hs10864043
Length=536
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AG+ VYK +P+G +E+ IPYL+RRA+EN +L GA+ E L E
Sbjct 463 QLLGMCDHVSLALGQAGYVVYKSIPYGSLEEVIPYLIRRAQENRSVLQGARREQELLSQE 522
Query 61 ITRRVL 66
+ RR+L
Sbjct 523 LWRRLL 528
> Hs19924111
Length=600
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
QL GM D S L AG+ VYKYVP+GPV + +PYL RRA EN+ ++ G E
Sbjct 526 QLLGMCDQISFPLGQAGYPVYKYVPYGPVMEVLPYLSRRALENSSLMKGTHRE 578
> HsM5174645
Length=516
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
QL GM D S L AG+ VYKYVP+GPV + +PYL RRA EN+ ++ G E
Sbjct 442 QLLGMCDQISFPLGQAGYPVYKYVPYGPVMEVLPYLSRRALENSSLMKGTHRE 494
> SPCC70.03c
Length=492
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 1 QLYGMGD------AFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEV 54
QL GM D A+SQ F + KYV GP+ + +PYL+RRAREN L + E
Sbjct 421 QLLGMADDITYALAYSQRNQQPNFCIVKYVSCGPISEVLPYLVRRARENIDALDRCKEER 480
Query 55 RYLCHEITRRVL 66
Y + RR+
Sbjct 481 AYYRQALRRRIF 492
> YLR142w
Length=476
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 20/45 (44%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 1 QLYGMGDAFSQGL--SSAGFDVYKYVPFGPVEDTIPYLLRRAREN 43
QL GM D + L + ++ KYVP+GP +T YLLRR +EN
Sbjct 406 QLLGMADNVTYDLITNHGAKNIIKYVPWGPPLETKDYLLRRLQEN 450
> YHL030w
Length=1868
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 10/32 (31%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 22 KYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
+Y+ P E+TI +L+ +A+E +LG ++
Sbjct 1660 RYINVNPQEETITFLIEKAKEMIRLLGSESDD 1691
Lambda K H
0.323 0.143 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200381448
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40