bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5603_orf1
Length=99
Score E
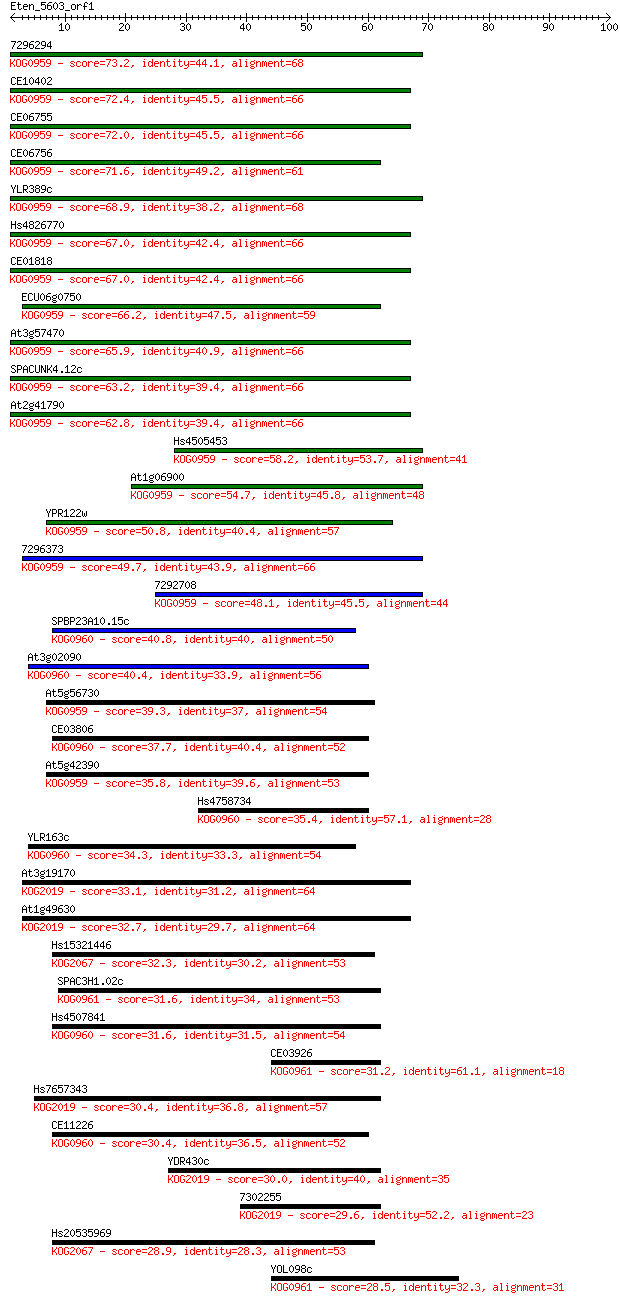
Sequences producing significant alignments: (Bits) Value
7296294 73.2 1e-13
CE10402 72.4 2e-13
CE06755 72.0 3e-13
CE06756 71.6 3e-13
YLR389c 68.9 2e-12
Hs4826770 67.0 7e-12
CE01818 67.0 9e-12
ECU06g0750 66.2 1e-11
At3g57470 65.9 2e-11
SPACUNK4.12c 63.2 1e-10
At2g41790 62.8 1e-10
Hs4505453 58.2 4e-09
At1g06900 54.7 4e-08
YPR122w 50.8 6e-07
7296373 49.7 1e-06
7292708 48.1 4e-06
SPBP23A10.15c 40.8 7e-04
At3g02090 40.4 8e-04
At5g56730 39.3 0.002
CE03806 37.7 0.005
At5g42390 35.8 0.022
Hs4758734 35.4 0.024
YLR163c 34.3 0.055
At3g19170 33.1 0.15
At1g49630 32.7 0.17
Hs15321446 32.3 0.19
SPAC3H1.02c 31.6 0.38
Hs4507841 31.6 0.40
CE03926 31.2 0.52
Hs7657343 30.4 0.91
CE11226 30.4 0.93
YDR430c 30.0 1.1
7302255 29.6 1.4
Hs20535969 28.9 2.6
YOL098c 28.5 2.9
> 7296294
Length=990
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
RD+R QL NG+ + + P ++ + A++ G + DP ++PGLAHF EHMLFLGT KY
Sbjct 35 RDYRGLQLENGLKVLLISDPNTDVSAAALSVQVGHMSDPTNLPGLAHFCEHMLFLGTEKY 94
Query 61 PEPESYDS 68
P Y +
Sbjct 95 PHENGYTT 102
> CE10402
Length=1067
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KY
Sbjct 83 REYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKY 142
Query 61 PEPESY 66
P Y
Sbjct 143 PSENEY 148
> CE06755
Length=980
Score = 72.0 bits (175), Expect = 3e-13, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KY
Sbjct 24 REYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKY 83
Query 61 PEPESY 66
P Y
Sbjct 84 PTENEY 89
> CE06756
Length=845
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 30/61 (49%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R+ R +L+NG+ + V P ++++ ++A G L DP ++PGLAHF EHMLFLGTSKY
Sbjct 24 RECRGLELTNGLRVLLVSDPTTDKSAVSLAVKAGHLMDPWELPGLAHFCEHMLFLGTSKY 83
Query 61 P 61
P
Sbjct 84 P 84
> YLR389c
Length=988
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 46/68 (67%), Gaps = 0/68 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R +R +L N + A+ + P++++A ++ N G+ DP+++PGLAHF EH+LF+G+ K+
Sbjct 72 RSYRFIELPNKLKALLIQDPKADKAAASLDVNIGAFEDPKNLPGLAHFCEHLLFMGSEKF 131
Query 61 PEPESYDS 68
P+ Y S
Sbjct 132 PDENEYSS 139
> Hs4826770
Length=1019
Score = 67.0 bits (162), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 45/66 (68%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R +L+NG+ + + P ++++ A+ + GSL DP ++ GL+HF EHMLFLGT KY
Sbjct 62 REYRGLELANGIKVLLMSDPTTDKSSAALDVHIGSLSDPPNIAGLSHFCEHMLFLGTKKY 121
Query 61 PEPESY 66
P+ Y
Sbjct 122 PKENEY 127
> CE01818
Length=745
Score = 67.0 bits (162), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
+ +R +L+NG+ + V ++ + A+ G L DP ++PGLAHF EHMLFLGT+KY
Sbjct 25 KKYRGLELTNGLRVLLVSDSKTRVSAVALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKY 84
Query 61 PEPESY 66
P Y
Sbjct 85 PSEREY 90
> ECU06g0750
Length=882
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 3 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 61
+ + ++ NGM A+ + P ++ AV+ GS DP D GLAHFLEHMLF+GT KYP
Sbjct 27 YEYVEIPNGMRALIMSDPGLDKCSCAVSVRVGSFDDPADAQGLAHFLEHMLFMGTEKYP 85
> At3g57470
Length=989
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R L N + + + P +++ ++ + GS DPE + GLAHFLEHMLF + KY
Sbjct 25 REYRRIVLKNSLEVLLISDPETDKCAASMNVSVGSFTDPEGLEGLAHFLEHMLFYASEKY 84
Query 61 PEPESY 66
PE +SY
Sbjct 85 PEEDSY 90
> SPACUNK4.12c
Length=969
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R +L N + + V P ++ A A+ + GS +P ++ GLAHF EH+LF+GT KY
Sbjct 22 REYRLIKLENDLEVLLVRDPETDNASAAIDVHIGSQSNPRELLGLAHFCEHLLFMGTKKY 81
Query 61 PEPESY 66
P+ Y
Sbjct 82 PDENEY 87
> At2g41790
Length=970
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
R++R L N + + + P +++ +++ + GS DP+ + GLAHFLEHMLF + KY
Sbjct 23 REYRMIVLKNLLQVLLISDPDTDKCAASMSVSVGSFSDPQGLEGLAHFLEHMLFYASEKY 82
Query 61 PEPESY 66
PE +SY
Sbjct 83 PEEDSY 88
> Hs4505453
Length=1219
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 22/41 (53%), Positives = 31/41 (75%), Gaps = 0/41 (0%)
Query 28 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDS 68
A+ GS DP+D+PGLAHFLEHM+F+G+ KYP+ +D+
Sbjct 282 ALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYPDENGFDA 322
> At1g06900
Length=1023
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 21 RSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDS 68
++ +A A+ + GS DP + GLAHFLEHMLF+G++++P+ YDS
Sbjct 103 QTKKAAAAMCVSMGSFLDPPEAQGLAHFLEHMLFMGSTEFPDENEYDS 150
> YPR122w
Length=1208
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 7 QLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLF-LGTSKYPEP 63
+L NG+ A+ + P + ++ TGS DP+D+ GLAH EHM+ G+ KYP+P
Sbjct 28 KLPNGILALIISDPTDTSSSCSLTVCTGSHNDPKDIAGLAHLCEHMILSAGSKKYPDP 85
> 7296373
Length=908
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 41/88 (46%), Gaps = 22/88 (25%)
Query 3 FRHYQLSNGMHAIAVH---------HPRSNE-------------AGFAVAANTGSLYDPE 40
+R LSNG+ A+ + H S E A AV GS +P+
Sbjct 50 YRALTLSNGLRAMLISDSYIDEPSIHRASRESLNSSTENFNGKLAACAVLVGVGSFSEPQ 109
Query 41 DVPGLAHFLEHMLFLGTSKYPEPESYDS 68
GLAHF+EHM+F+G+ K+P +DS
Sbjct 110 QYQGLAHFVEHMIFMGSEKFPVENEFDS 137
> 7292708
Length=1077
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 25 AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDS 68
A A+ + GS +P GLAHFLEHM+F+G+ KYP+ +D+
Sbjct 94 AACALLIDYGSFAEPTKYQGLAHFLEHMIFMGSEKYPKENIFDA 137
> SPBP23A10.15c
Length=457
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGT 57
L NG+ HHP + A V + GS + G AHFLEH+ F GT
Sbjct 27 LKNGLTVATEHHPYAQTATVLVGVDAGSRAETAKNNGAAHFLEHLAFKGT 76
> At3g02090
Length=531
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 4 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 59
R L NG+ + + A V + GS ++ ++ G AHFLEHM+F GT +
Sbjct 98 RVTTLPNGLRVATESNLSAKTATVGVWIDAGSRFESDETNGTAHFLEHMIFKGTDR 153
> At5g56730
Length=956
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 9/59 (15%)
Query 7 QLSNGMHAIAVHHPRSN-----EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
+L NG+ +++ R N A A+A GS+ + ED G+AH +EH+ F T++Y
Sbjct 44 RLDNGL----IYYVRRNSKPRMRAALALAVKVGSVLEEEDQRGVAHIVEHLAFSATTRY 98
> CE03806
Length=485
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 59
L NG +A + + A V + GS Y+ E G AHFLEHM F GT +
Sbjct 62 LPNGFR-VATENTGGSTATIGVFIDAGSRYENEKNNGTAHFLEHMAFKGTPR 112
> At5g42390
Length=1265
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 5/56 (8%)
Query 7 QLSNGMHAIAVHH---PRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 59
QL NG+ + + + P EA V + GS+ + ED G+AH +EH+ FLG+ K
Sbjct 199 QLKNGLRYLILPNKVPPNRFEAHMEV--HVGSIDEEEDEQGIAHMIEHVAFLGSKK 252
> Hs4758734
Length=489
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 32 NTGSLYDPEDVPGLAHFLEHMLFLGTSK 59
+ GS Y+ E G AHFLEHM F GT K
Sbjct 86 DAGSRYENEKNNGTAHFLEHMAFKGTKK 113
> YLR163c
Length=462
Score = 34.3 bits (77), Expect = 0.055, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 4 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGT 57
R +L NG+ + P ++ A + + GS + G AHFLEH+ F GT
Sbjct 27 RTSKLPNGLTIATEYIPNTSSATVGIFVDAGSRAENVKNNGTAHFLEHLAFKGT 80
> At3g19170
Length=1052
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 7/64 (10%)
Query 3 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 62
F+H + G ++V + N+ F V T P+D G+ H LEH + G+ KYP
Sbjct 97 FKHKK--TGCEVMSVSNEDENKV-FGVVFRT----PPKDSTGIPHILEHSVLCGSRKYPV 149
Query 63 PESY 66
E +
Sbjct 150 KEPF 153
> At1g49630
Length=1076
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 7/64 (10%)
Query 3 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 62
F+H + G ++V + N+ F + T P+D G+ H LEH + G+ KYP
Sbjct 124 FKHKK--TGCEVMSVSNDDENKV-FGIVFRT----PPKDSTGIPHILEHSVLCGSRKYPM 176
Query 63 PESY 66
E +
Sbjct 177 KEPF 180
> Hs15321446
Length=525
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
L NG+ +A + + N+GS Y+ + + G+AHFLE + F T+++
Sbjct 72 LDNGLR-VASQNKFGQFCTVGILINSGSRYEAKYLSGIAHFLEKLAFSSTARF 123
> SPAC3H1.02c
Length=1036
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 5/53 (9%)
Query 9 SNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 61
+ G +V P S G V A + D G H LEH+ F+G+ KYP
Sbjct 28 ATGFSVASVKTPTSRLQGSFVVAT-----EAHDNLGCPHTLEHLCFMGSKKYP 75
> Hs4507841
Length=480
Score = 31.6 bits (70), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 24/54 (44%), Gaps = 1/54 (1%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 61
L NG+ +A V + GS ++ E G +FLEH+ F GT P
Sbjct 53 LDNGLR-VASEQSSQPTCTVGVWIDVGSRFETEKNNGAGYFLEHLAFKGTKNRP 105
> CE03926
Length=995
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 44 GLAHFLEHMLFLGTSKYP 61
GL H LEH++F+G+ KYP
Sbjct 57 GLPHTLEHLVFMGSKKYP 74
> Hs7657343
Length=1038
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 30/80 (37%), Gaps = 23/80 (28%)
Query 5 HYQLSNGMHAIAVHHPRSNEAGFAVAA-----NTGSLY------------------DPED 41
Y+L + +H V+ S F A +TG+ Y P D
Sbjct 39 QYKLGDKIHGFTVNQVTSVPELFLTAVKLTHDDTGARYLHLAREDTNNLFSVQFRTTPMD 98
Query 42 VPGLAHFLEHMLFLGTSKYP 61
G+ H LEH + G+ KYP
Sbjct 99 STGVPHILEHTVLCGSQKYP 118
> CE11226
Length=471
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 26/52 (50%), Gaps = 1/52 (1%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 59
L NG + + S A V TGS ++ E G+AHFLE ++ GT K
Sbjct 43 LKNGFRVVTEDNG-SATATVGVWIETGSRFENEKNNGVAHFLERLIHKGTGK 93
> YDR430c
Length=989
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 4/35 (11%)
Query 27 FAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 61
F++A T +P D G+ H LEH G+ KYP
Sbjct 68 FSIAFKT----NPPDSTGVPHILEHTTLCGSVKYP 98
> 7302255
Length=1112
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 39 PEDVPGLAHFLEHMLFLGTSKYP 61
P D GL H LEH+ G+ KYP
Sbjct 120 PFDSTGLPHILEHLSLCGSQKYP 142
> Hs20535969
Length=417
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 1/53 (1%)
Query 8 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 60
L NG +A + + N+GS Y+ + G+AHFLE + F +++
Sbjct 20 LDNGFR-VASQNKFGQFCIVGILINSGSRYEATYLSGIAHFLEKLAFSSNARF 71
> YOL098c
Length=1037
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 10/31 (32%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 44 GLAHFLEHMLFLGTSKYPEPESYDSSSAVAI 74
G H LEH++F+G+ YP D++ +++
Sbjct 57 GAPHTLEHLIFMGSKSYPYKGLLDTAGNLSL 87
Lambda K H
0.319 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40