bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5538_orf2
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
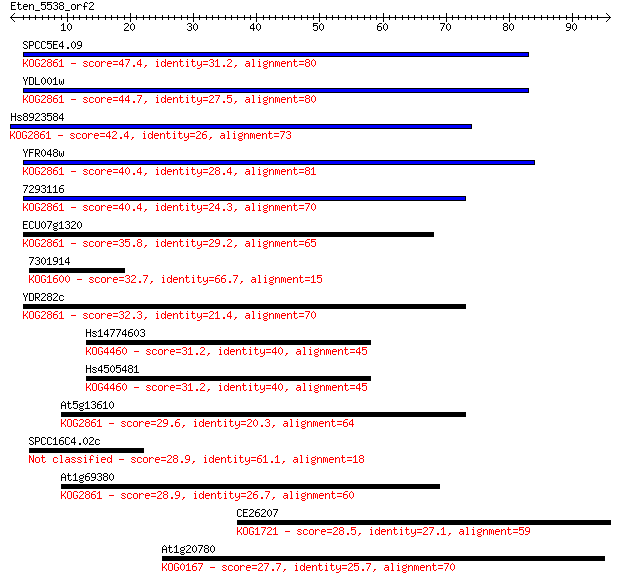
SPCC5E4.09 47.4 7e-06
YDL001w 44.7 5e-05
Hs8923584 42.4 2e-04
YFR048w 40.4 8e-04
7293116 40.4 9e-04
ECU07g1320 35.8 0.021
7301914 32.7 0.18
YDR282c 32.3 0.23
Hs14774603 31.2 0.48
Hs4505481 31.2 0.51
At5g13610 29.6 1.4
SPCC16C4.02c 28.9 2.2
At1g69380 28.9 2.6
CE26207 28.5 2.9
At1g20780 27.7 4.7
> SPCC5E4.09
Length=446
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 3 DVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTW 62
D P+ W + + + + +LEI +R+ LLN R I +LL +++E+ + L W
Sbjct 298 DSPELMWTEPQLEPIYTAARSYLEINQRVALLNQRVEVIGDLLSMLKEQITHTHDESLEW 357
Query 63 IIVILLFFQVVALALRNFVI 82
I+VIL+ +V +AL + V+
Sbjct 358 IVVILMGL-LVLIALFSIVV 376
> YDL001w
Length=430
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Query 3 DVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTW 62
D P+ W++ + + ++ +LEI +R+ LLN R I +LL++++E+ E L +
Sbjct 345 DSPEIMWSEPQLEPIYQATRGYLEINQRVSLLNQRLEVISDLLQMLKEQLGHSHEEYLEF 404
Query 63 IIVILLFFQVVALALRNFVI 82
I+++L+ +V+ +++ N V+
Sbjct 405 IVILLVGVEVL-ISVINIVV 423
> Hs8923584
Length=449
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 1 FRDVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRL 60
F P ++W+ + + + + L I +R+K++N + EL ++R ++ RL
Sbjct 369 FLITPDFYWDRENLEGLYDKTCQFLSIGRRVKVMNEKLQHCMELTDLMRNHLNEKRALRL 428
Query 61 TWIIVILLFFQVV 73
W+IVIL+ +V+
Sbjct 429 EWMIVILITIEVM 441
> YFR048w
Length=662
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 45/86 (52%), Gaps = 5/86 (5%)
Query 3 DVPQYFWN-DDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLT 61
D P++FW+ + + + +LEI +R+++LN+R E + + R R+T
Sbjct 572 DTPEFFWSFEPSLHPLYVAMREYLEIDQRVQVLNDRCKVFLEFFDICVDSVAERNMARVT 631
Query 62 -WIIVILLF---FQVVALALRNFVIH 83
W I+++LF F + + +R +IH
Sbjct 632 WWFILVILFGVIFSLTEIFVRYVIIH 657
> 7293116
Length=420
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 3 DVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTW 62
D P ++W+ + + + ++ + I +R K++N + EL ++V RL W
Sbjct 320 DAPDFYWDREELEALYLQVCSYFSISRRTKVMNEKINHCVELAELVSHNLNDAHHIRLEW 379
Query 63 IIVILLFFQV 72
+I+IL+ +V
Sbjct 380 MIIILIMVEV 389
> ECU07g1320
Length=334
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 38/67 (56%), Gaps = 2/67 (2%)
Query 3 DVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVRE--ERRSRQEFRL 60
D P++ W+ + + +L+I+ R +LLN R I+ +L+++ E R S ++F +
Sbjct 249 DEPEFVWDYPAFSSLYETCKRYLDIKPRAELLNRRCDVINGILEILNENTNRSSIEKFEV 308
Query 61 TWIIVIL 67
I++IL
Sbjct 309 VLIVIIL 315
> 7301914
Length=480
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 10/15 (66%), Positives = 11/15 (73%), Gaps = 0/15 (0%)
Query 4 VPQYFWNDDRWQYFW 18
VP YFWN+D W FW
Sbjct 323 VPWYFWNEDLWMSFW 337
> YDR282c
Length=414
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 15/70 (21%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 3 DVPQYFWNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTW 62
+ P +W++ + + ++ + +L+I RI +LN++ + + + R L W
Sbjct 318 ETPDLYWSEPQLEEIFKNVSRYLDIGPRINILNSKLDYSTDECRALISLLNERNSTFLEW 377
Query 63 IIVILLFFQV 72
II+ L+ F++
Sbjct 378 IIIYLIAFEL 387
> Hs14774603
Length=741
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 13 RWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQE 57
R QY ++ EIQ+R+KLL ++ E L REER+S +E
Sbjct 573 REQYILKQDLAKEEIQRRVKLLCDQKKKQLEDLSYCREERKSLRE 617
> Hs4505481
Length=741
Score = 31.2 bits (69), Expect = 0.51, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 13 RWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQE 57
R QY ++ EIQ+R+KLL ++ E L REER+S +E
Sbjct 573 REQYILKQDLAKEEIQRRVKLLCDQKKKQLEDLSYCREERKSLRE 617
> At5g13610
Length=402
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 13/64 (20%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 9 WNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTWIIVILL 68
W D ++ W L E+ + L+ + + ++ ++E ++R+ L W+I+IL+
Sbjct 327 WKDAKYGQIWEFLRDEFELTQSFANLDYKLKFVEHNVRFLQEILQNRKSATLEWLIIILI 386
Query 69 FFQV 72
++
Sbjct 387 SMEI 390
> SPCC16C4.02c
Length=548
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 1/19 (5%)
Query 4 VPQYFWNDDRWQY-FWRRL 21
VPQ WNDD WQ +W+ L
Sbjct 525 VPQSIWNDDIWQEPYWKNL 543
> At1g69380
Length=373
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 28/60 (46%), Gaps = 0/60 (0%)
Query 9 WNDDRWQYFWRRLYVHLEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTWIIVILL 68
W + R+ + L EI +R L+ + I + ++E ++RQ L W I+ LL
Sbjct 294 WREARYAQIYEYLREEYEISQRFGDLDYKLKFIEHNIHFLQEVMQNRQSDLLEWCIIFLL 353
> CE26207
Length=364
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query 37 RYFCIHELLKVVREERRSRQEFRLTWIIVILLFFQVVALALRNFVIHPYEQIDGKSRRG 95
RY + E+ KV E R++ + F + I ++ F +AL+N+ + + +I +RG
Sbjct 188 RYMVLDEMEKVA-ENRKTHEFFEVNVISSVIFFVNFDKIALKNYFCYIFSKIGIGFQRG 245
> At1g20780
Length=801
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 18/76 (23%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 25 LEIQKRIKLLNNRYFCIHELLKVVREERRSRQEFRLTWIIVILLFFQVVA------LALR 78
L+++K +K+L+ I +L V+RE R R R W++ +L + +A +L
Sbjct 704 LDVEKGVKILDEAD-GIRHILNVLRENRTERLTRRAVWMVERILRIEDIAREVAEEQSLS 762
Query 79 NFVIHPYEQIDGKSRR 94
++ ++ D ++R+
Sbjct 763 AALVDAFQNADFRTRQ 778
Lambda K H
0.335 0.148 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40