bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5459_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
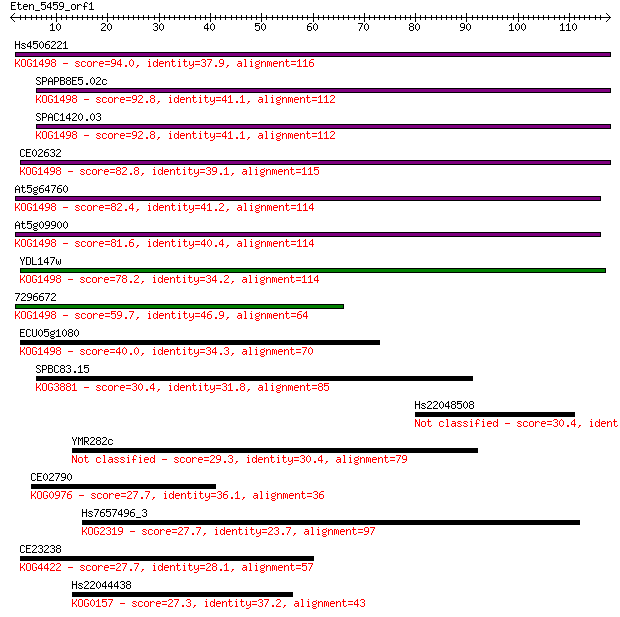
Hs4506221 94.0 6e-20
SPAPB8E5.02c 92.8 1e-19
SPAC1420.03 92.8 1e-19
CE02632 82.8 1e-16
At5g64760 82.4 2e-16
At5g09900 81.6 4e-16
YDL147w 78.2 3e-15
7296672 59.7 1e-09
ECU05g1080 40.0 0.001
SPBC83.15 30.4 0.74
Hs22048508 30.4 0.93
YMR282c 29.3 1.9
CE02790 27.7 5.3
Hs7657496_3 27.7 5.6
CE23238 27.7 5.7
Hs22044438 27.3 6.4
> Hs4506221
Length=456
Score = 94.0 bits (232), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 78/116 (67%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M+++E+ ++IL+QM L L D+IR QI+SKK++ K + + + LK+KY
Sbjct 167 QELQVETYGSMEKKERVEFILEQMRLCLAVKDYIRTQIISKKINTKFFQEENTEKLKLKY 226
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM HE + L + K + IY+T I+ + + + L+ ++++LAPFD+E
Sbjct 227 YNLMIQLDQHEGSYLSICKHYRAIYDTPCIQAESEKWQQALKSVVLYVILAPFDNE 282
> SPAPB8E5.02c
Length=443
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/112 (41%), Positives = 70/112 (62%), Gaps = 0/112 (0%)
Query 6 VETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYYELM 65
VET+G+ D +EK +IL Q+ L L +SD+ +KK++ K E D+Q LK+KYYE
Sbjct 159 VETYGSFDLKEKVAFILDQVRLFLLRSDYYMASTYTKKINVKFFEKEDVQSLKLKYYEQK 218
Query 66 TVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
LH+DA LDV K + +Y+T ++++DP + E LE F +L P+D+E
Sbjct 219 IRIGLHDDAYLDVCKYYRAVYDTAVVQEDPEKWKEILENVVCFALLTPYDNE 270
> SPAC1420.03
Length=443
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/112 (41%), Positives = 70/112 (62%), Gaps = 0/112 (0%)
Query 6 VETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYYELM 65
VET+G+ D +EK +IL Q+ L L +SD+ +KK++ K E D+Q LK+KYYE
Sbjct 159 VETYGSFDLKEKVAFILDQVRLFLLRSDYYMASTYTKKINVKFFEKEDVQSLKLKYYEQK 218
Query 66 TVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
LH+DA LDV K + +Y+T ++++DP + E LE F +L P+D+E
Sbjct 219 IRIGLHDDAYLDVCKYYRAVYDTAVVQEDPEKWKEILENVVCFALLTPYDNE 270
> CE02632
Length=490
Score = 82.8 bits (203), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 72/118 (61%), Gaps = 3/118 (2%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAAD---LQVLKV 59
++QVET+G+M+ EK +Y+L+QM L ++DF+R I+SKK++ K +D +Q LK+
Sbjct 190 ELQVETYGSMEMREKVQYLLEQMRYSLVRNDFVRATIISKKINIKFFNKSDEDEVQNLKL 249
Query 60 KYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
KYY+ M LH+ LDV + IY T+ I+ D A+ L ++ +LAP +E
Sbjct 250 KYYDSMIRIGLHDGNYLDVCRHHREIYETKKIKADSAKATSHLRSAIVYCLLAPHTNE 307
> At5g64760
Length=529
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/137 (34%), Positives = 75/137 (54%), Gaps = 23/137 (16%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA---------- 51
++V VETFGAM + EK +IL+Q+ L L + DF+R QI+S+K++P++ +A
Sbjct 221 QEVAVETFGAMAKTEKIAFILEQVRLCLDQKDFVRAQILSRKINPRVFDADTTKEKKKPK 280
Query 52 ----------AD---LQVLKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARM 98
AD L VLK YYELM YY H + +++ + + IY+ ++++P +
Sbjct 281 EGENMVEEAPADIPTLLVLKRIYYELMIRYYSHNNEYIEICRSYKAIYDIPSVKENPEQW 340
Query 99 NECLEFFSIFMVLAPFD 115
L F+ LAP D
Sbjct 341 IPVLRKICWFLALAPHD 357
> At5g09900
Length=442
Score = 81.6 bits (200), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 74/137 (54%), Gaps = 23/137 (16%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA---------- 51
++V VETFGAM + EK +IL+Q+ L L + DF+R QI+S+K++P++ +A
Sbjct 134 QEVAVETFGAMAKTEKIAFILEQVRLCLDRQDFVRAQILSRKINPRVFDADTKKDKKKPK 193
Query 52 ----------ADLQV---LKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARM 98
AD+ LK YYELM YY H + +++ + + IY+ +++ P +
Sbjct 194 EGDNMVEEAPADIPTLLELKRIYYELMIRYYSHNNEYIEICRSYKAIYDIPSVKETPEQW 253
Query 99 NECLEFFSIFMVLAPFD 115
L F+VLAP D
Sbjct 254 IPVLRKICWFLVLAPHD 270
> YDL147w
Length=445
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 68/114 (59%), Gaps = 0/114 (0%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYY 62
++QVET+G+M+ EK ++IL+QM L + K D+ + ++S+K+ K + + LK++YY
Sbjct 162 ELQVETYGSMEMSEKIQFILEQMELSILKGDYSQATVLSRKILKKTFKNPKYESLKLEYY 221
Query 63 ELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDS 116
L+ LH+ L+VA+ IY T I+ D A+ L F+VL+P+ +
Sbjct 222 NLLVKISLHKREYLEVAQYLQEIYQTDAIKSDEAKWKPVLSHIVYFLVLSPYGN 275
> 7296672
Length=502
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 45/64 (70%), Gaps = 0/64 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
E++QVET+G+MD+ EK + IL+QM L L K D++ QI++KK+S K + LK+K+
Sbjct 167 EELQVETYGSMDKREKVELILEQMRLCLLKEDYVSTQIIAKKISIKFFDDPAQHDLKLKF 226
Query 62 YELM 65
Y LM
Sbjct 227 YYLM 230
> ECU05g1080
Length=387
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYY 62
+V VETF + Y L+Q+ L + D+IR I KK+ K E D K+K+Y
Sbjct 120 NVPVETFTMVKESVVVNYQLEQLRLCVSNLDWIRADITMKKIRKKYFEENDAAEEKIKFY 179
Query 63 ELMTVYYLHE 72
EL+ +L +
Sbjct 180 ELVVQLHLGQ 189
> SPBC83.15
Length=387
Score = 30.4 bits (67), Expect = 0.74, Method: Composition-based stats.
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 18/96 (18%)
Query 6 VETFGAMDREEKTKYILKQMM--LLLRKSDFIRCQIVSK---------KLSPKLLEAADL 54
++ FG +DRE+ ++LK M + RK+ I C V++ +L LLE A +
Sbjct 33 IQKFGELDREKGVLFMLKHEMNVFVARKNGTIECWNVNQEPPILSSLWQLDSSLLETASI 92
Query 55 QVLKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQI 90
+K LM AL D F HI ++++
Sbjct 93 VSMKYSNGWLML-------ALSDGNLLFRHIESSKL 121
> Hs22048508
Length=99
Score = 30.4 bits (67), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 80 KCFGHIYNTQIIRDDPARMNECLEFFSIFMV 110
KC G ++N I D+ M+EC SIF++
Sbjct 65 KCMGDLWNASICGDEHVTMHECETVLSIFIL 95
> YMR282c
Length=580
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Query 13 DREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPK-LLEAADLQVLKVKYYELMTVYYLH 71
D E+ + L++M+ L K+ IR K + + LL+ ADL + + K
Sbjct 264 DNSEQKQGFLRKMVRLGAKNTSIRLSSTYKAMDHQTLLKIADLALQEKKLLN-------S 316
Query 72 EDALLDVAKCFGHIYNTQII 91
ED L + + FGH+ TQI+
Sbjct 317 EDLLSTLIQSFGHLGQTQIL 336
> CE02790
Length=1286
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 5 QVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIV 40
Q T D E+ TKY+ ++ M L R+ IRC+++
Sbjct 313 QTTTASLGDSEQATKYLHEENMKLTRQKADIRCELL 348
> Hs7657496_3
Length=379
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 49/98 (50%), Gaps = 5/98 (5%)
Query 15 EEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYYELMTVYYLHEDA 74
+EK +++ + LR ++ Q++ ++ + E +D+ V + M + D
Sbjct 119 DEKKDLAIQKRIRALR---WVTPQMLCVPVNEDIPEVSDMVVKAITDIIEMDSKRVPRDK 175
Query 75 LLDVAKCFGHIYNT-QIIRDDPARMNECLEFFSIFMVL 111
L + KC HI+N +I +++PA ++ L I++VL
Sbjct 176 LACITKCSKHIFNAIKITKNEPASADDFLPTL-IYIVL 212
> CE23238
Length=660
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFI--RCQIVSKKLSPKLLEAADLQVLKV 59
+VQ+ T G +RE+ T + K + + + S F R + + +KLSP + L + ++
Sbjct 503 NVQLHTLGTSEREQFTSLVQKLVAVWVEFSQFTEERMRRLQRKLSPSQISECALLLTRI 561
> Hs22044438
Length=526
Score = 27.3 bits (59), Expect = 6.4, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 13 DREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQ 55
DR+E K LKQ R+ DF+ + +K + K ADLQ
Sbjct 449 DRKESLKDKLKQDTTQKRRQDFLDILLSAKSENTKDFSEADLQ 491
Lambda K H
0.326 0.140 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40