bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
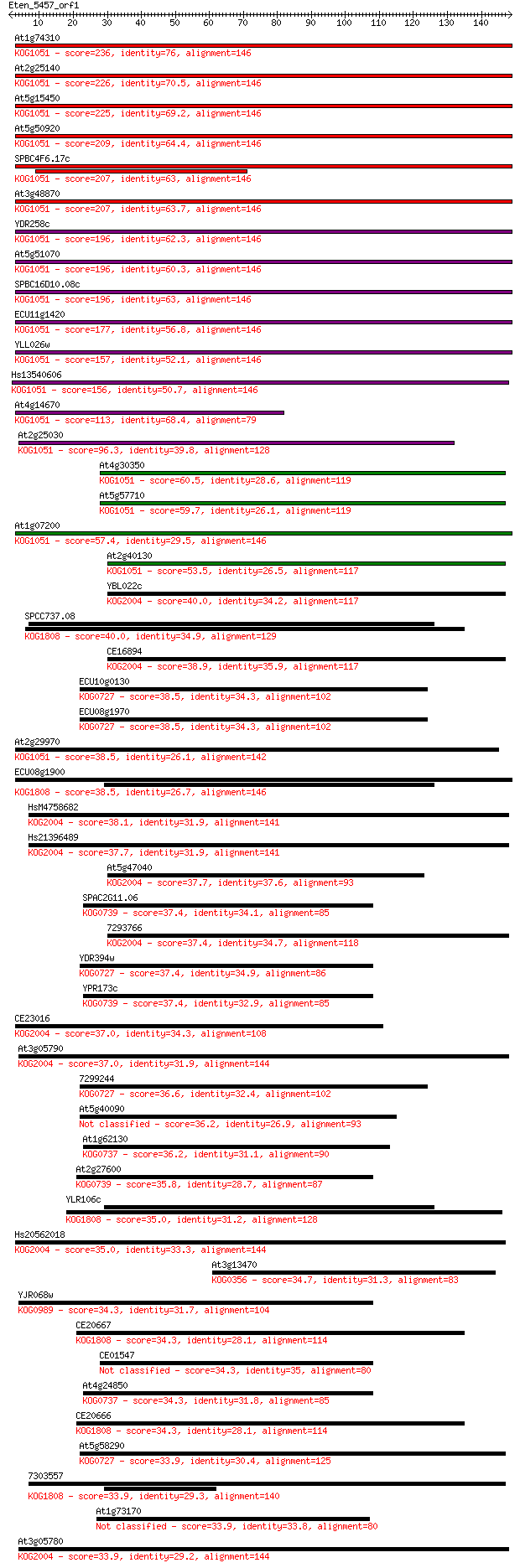
Query= Eten_5457_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
At1g74310 236 2e-62
At2g25140 226 9e-60
At5g15450 225 2e-59
At5g50920 209 1e-54
SPBC4F6.17c 207 4e-54
At3g48870 207 5e-54
YDR258c 196 1e-50
At5g51070 196 1e-50
SPBC16D10.08c 196 1e-50
ECU11g1420 177 5e-45
YLL026w 157 9e-39
Hs13540606 156 2e-38
At4g14670 113 1e-25
At2g25030 96.3 2e-20
At4g30350 60.5 1e-09
At5g57710 59.7 2e-09
At1g07200 57.4 1e-08
At2g40130 53.5 1e-07
YBL022c 40.0 0.002
SPCC737.08 40.0 0.002
CE16894 38.9 0.004
ECU10g0130 38.5 0.004
ECU08g1970 38.5 0.004
At2g29970 38.5 0.004
ECU08g1900 38.5 0.005
HsM4758682 38.1 0.007
Hs21396489 37.7 0.007
At5g47040 37.7 0.008
SPAC2G11.06 37.4 0.010
7293766 37.4 0.011
YDR394w 37.4 0.012
YPR173c 37.4 0.012
CE23016 37.0 0.013
At3g05790 37.0 0.014
7299244 36.6 0.018
At5g40090 36.2 0.022
At1g62130 36.2 0.022
At2g27600 35.8 0.032
YLR106c 35.0 0.046
Hs20562018 35.0 0.050
At3g13470 34.7 0.065
YJR068w 34.3 0.078
CE20667 34.3 0.079
CE01547 34.3 0.084
At4g24850 34.3 0.093
CE20666 34.3 0.099
At5g58290 33.9 0.10
7303557 33.9 0.11
At1g73170 33.9 0.11
At3g05780 33.9 0.12
> At1g74310
Length=911
Score = 236 bits (601), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 111/146 (76%), Positives = 126/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AVSE+ILRS AGL R +P GSFLFLGPTGVGKTEL KA+AE+LFD + +VR+DMS
Sbjct 577 AVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMS 636
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RLIGAPPGYVGHEEGGQLTEAVRR PY V+L DEVEKAH V+N LLQVLD
Sbjct 637 EYMEQHSVSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLD 696
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTD QGRTVDF N ++I+TSN+G
Sbjct 697 DGRLTDGQGRTVDFRNSVIIMTSNLG 722
> At2g25140
Length=874
Score = 226 bits (577), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 103/146 (70%), Positives = 127/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV +V+++I RS AGL+ N+PI SF+F+GPTGVGKTEL KA+A LF+T+ IVR+DMS
Sbjct 571 AVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMS 630
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RL+GAPPGYVG+EEGGQLTE VRR PYSVVL DE+EKAH V+N+LLQ+LD
Sbjct 631 EYMEKHSVSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLD 690
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+TDSQGRTV F NC++I+TSNIG
Sbjct 691 DGRITDSQGRTVSFKNCVVIMTSNIG 716
> At5g15450
Length=968
Score = 225 bits (574), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 101/146 (69%), Positives = 127/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AVTAV+E+I RS AGL+ +PI SF+F+GPTGVGKTEL KA+A +F+T+E +VR+DMS
Sbjct 656 AVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMS 715
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME H+V+RLIGAPPGYVG+EEGGQLTE VRR PYSV+L DE+EKAH V+NV LQ+LD
Sbjct 716 EYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQILD 775
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+TDSQGRTV F+N ++I+TSN+G
Sbjct 776 DGRVTDSQGRTVSFTNTVIIMTSNVG 801
> At5g50920
Length=929
Score = 209 bits (533), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 122/146 (83%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S +I R+ GL N+PI SF+F GPTGVGK+EL KA+A F ++E ++RLDMS
Sbjct 616 AVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMS 675
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ME H+V++LIG+PPGYVG+ EGGQLTEAVRR PY+VVL DE+EKAH V+N++LQ+L+
Sbjct 676 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILE 735
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS+GRTVDF N LLI+TSN+G
Sbjct 736 DGRLTDSKGRTVDFKNTLLIMTSNVG 761
> SPBC4F6.17c
Length=803
Score = 207 bits (528), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 92/146 (63%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ A+++++ S AGL N+P+ SFLFLGPTGVGKT L KA+AE LFDT + ++R DMS
Sbjct 510 ALKAIADAVRLSRAGLQNTNRPLASFLFLGPTGVGKTALTKALAEFLFDTDKAMIRFDMS 569
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ E H++ARLIG+PPGY+G+EE G+LTEAVRR PY+V+L DE+EKAHH + N+LLQVLD
Sbjct 570 EFQEKHTIARLIGSPPGYIGYEESGELTEAVRRKPYAVLLFDELEKAHHDITNLLLQVLD 629
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EG LTDSQGR VDF + L+++TSN+G
Sbjct 630 EGFLTDSQGRKVDFRSTLIVMTSNLG 655
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 32/62 (51%), Gaps = 9/62 (14%)
Query 9 ESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESH 68
E I R+ L+RR K + +GP GVGKT + + +A RI+R ++ E M+
Sbjct 118 EEIQRTIQILSRRTK--NNPALVGPAGVGKTAIMEGLA-------SRIIRGEVPESMKDK 168
Query 69 SV 70
V
Sbjct 169 RV 170
> At3g48870
Length=952
Score = 207 bits (528), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 93/146 (63%), Positives = 122/146 (83%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S +I R+ GL N+PI SF+F GPTGVGK+EL KA+A F ++E ++RLDMS
Sbjct 637 AVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMS 696
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ME H+V++LIG+PPGYVG+ EGGQLTEAVRR PY++VL DE+EKAH V+N++LQ+L+
Sbjct 697 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQILE 756
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS+GRTVDF N LLI+TSN+G
Sbjct 757 DGRLTDSKGRTVDFKNTLLIMTSNVG 782
> YDR258c
Length=811
Score = 196 bits (499), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 91/146 (62%), Positives = 112/146 (76%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ A+S+++ AGL +PI SF+FLGPTG GKTEL KA+AE LFD + ++R DMS
Sbjct 512 AIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNVIRFDMS 571
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ E H+V+RLIGAPPGYV E GGQLTEAVRR PY+VVL DE EKAH V +LLQVLD
Sbjct 572 EFQEKHTVSRLIGAPPGYVLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVSKLLLQVLD 631
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EG+LTDS G VDF N ++++TSNIG
Sbjct 632 EGKLTDSLGHHVDFRNTIIVMTSNIG 657
> At5g51070
Length=945
Score = 196 bits (499), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 88/146 (60%), Positives = 116/146 (79%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S ++ RS GL ++PI + LF GPTGVGKTEL KA+A F ++E ++RLDMS
Sbjct 635 AVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSEESMLRLDMS 694
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME H+V++LIG+PPGYVG EEGG LTEA+RR P++VVL DE+EKAH ++N+LLQ+ +
Sbjct 695 EYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPDIFNILLQLFE 754
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+G LTDSQGR V F N L+I+TSN+G
Sbjct 755 DGHLTDSQGRRVSFKNALIIMTSNVG 780
> SPBC16D10.08c
Length=905
Score = 196 bits (499), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 92/146 (63%), Positives = 115/146 (78%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AVTAV+ +I S AGL+ N+PI SFLF GP+G GKT L KA+A +FD + ++R+DMS
Sbjct 592 AVTAVANAIRLSRAGLSDPNQPIASFLFCGPSGTGKTLLTKALASFMFDDENAMIRIDMS 651
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RLIGAPPGYVGHE GGQLTE +RR PYSV+L DE+EKA +V VLLQVLD
Sbjct 652 EYMEKHSVSRLIGAPPGYVGHEAGGQLTEQLRRRPYSVILFDEIEKAAPEVLTVLLQVLD 711
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+T QG+ VD N ++I+TSN+G
Sbjct 712 DGRITSGQGQVVDAKNAVIIMTSNLG 737
> ECU11g1420
Length=851
Score = 177 bits (450), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 83/146 (56%), Positives = 110/146 (75%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+ +SIL+S GL ++P+GSFL LGPTGVGKTEL KAVA ELFD ++ ++ +DMS
Sbjct 560 AVDAIVDSILQSRVGLDDDDRPVGSFLLLGPTGVGKTELAKAVAMELFDNEKDMLVIDMS 619
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EY + +LIGA GYVG+ +GG LTE ++ PY+V+LLDEV+ AH V N L Q+LD
Sbjct 620 EYGNEMGITKLIGANAGYVGYNQGGTLTEPIKGRPYNVILLDEVDLAHQTVLNTLYQLLD 679
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EGR+TD +G VDF NC++I+TSN+G
Sbjct 680 EGRVTDGKGAVVDFRNCVVIMTSNLG 705
> YLL026w
Length=908
Score = 157 bits (396), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 76/146 (52%), Positives = 108/146 (73%), Gaps = 1/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ AVS ++ S +GLA +P SFLFLG +G GKTEL K VA LF+ ++ ++R+D S
Sbjct 586 AIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCS 644
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E E ++V++L+G GYVG++EGG LT ++ PYSV+L DEVEKAH V V+LQ+LD
Sbjct 645 ELSEKYAVSKLLGTTAGYVGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVMLQMLD 704
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+T QG+T+D SNC++I+TSN+G
Sbjct 705 DGRITSGQGKTIDCSNCIVIMTSNLG 730
> Hs13540606
Length=707
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 74/147 (50%), Positives = 103/147 (70%), Gaps = 2/147 (1%)
Query 2 AAVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELF-DTKERIVRLD 60
+A+ V +I R G P+ FLFLG +G+GKTEL K A+ + D K+ +RLD
Sbjct 352 SAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRLD 410
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
MSE+ E H VA+ IG+PPGYVGHEEGGQLT+ +++ P +VVL DEV+KAH V ++LQ+
Sbjct 411 MSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIMLQL 470
Query 121 LDEGRLTDSQGRTVDFSNCLLILTSNI 147
DEGRLTD +G+T+D + + I+TSN+
Sbjct 471 FDEGRLTDGKGKTIDCKDAIFIMTSNV 497
> At4g14670
Length=623
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 54/79 (68%), Positives = 64/79 (81%), Gaps = 0/79 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AV+ +ILRS GL R +P GSFLFLGPTGVGKTEL KA+AE+LFD++ +VRLDMS
Sbjct 542 AVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMS 601
Query 63 EYMESHSVARLIGAPPGYV 81
EY + SV +LIGAPPGYV
Sbjct 602 EYNDKFSVNKLIGAPPGYV 620
> At2g25030
Length=265
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/128 (39%), Positives = 74/128 (57%), Gaps = 33/128 (25%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSE 63
V +V+++I S AG++ N+ I SF+F+G V
Sbjct 17 VESVADAIRCSKAGISDPNRLIASFMFMGQPSV--------------------------- 49
Query 64 YMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDE 123
V++L+GA PGYVG+ +GG+LTE VRR PYSVV DE+EK H V+++LLQ+LD+
Sbjct 50 ------VSQLVGASPGYVGYGDGGKLTEVVRRRPYSVVQFDEIEKPHLDVFSILLQLLDD 103
Query 124 GRLTDSQG 131
GR+T+S G
Sbjct 104 GRITNSHG 111
> At4g30350
Length=924
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 64/119 (53%), Gaps = 9/119 (7%)
Query 28 FLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG 87
+F GP GK+++ A+++ + ++ + L S M+ R G
Sbjct 611 LMFTGPDRAGKSKMASALSDLVSGSQPITISLGSSSRMDDGLNIR---------GKTALD 661
Query 88 QLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLILTSN 146
+ EAVRRNP++V++L+++++A + N + ++ GR+ DS GR V N ++ILT+N
Sbjct 662 RFAEAVRRNPFAVIVLEDIDEADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
> At5g57710
Length=990
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 64/119 (53%), Gaps = 8/119 (6%)
Query 28 FLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG 87
LF GP VGK ++ A++ ++ T +++L + + + + G
Sbjct 655 LLFSGPDRVGKRKMVSALSSLVYGTNPIMIQLGSRQDAGDGNSS--------FRGKTALD 706
Query 88 QLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLILTSN 146
++ E V+R+P+SV+LL+++++A V + Q +D GR+ DS GR + N + ++T++
Sbjct 707 KIAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
> At1g07200
Length=979
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 67/147 (45%), Gaps = 9/147 (6%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDM 61
AV A+S+ I RRN+ G +L LGP VGK ++ ++E F K + +D
Sbjct 635 AVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSEVFFGGKVNYICVDF 694
Query 62 SEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVL 121
A + G +T + R P+SVVLL+ VEKA L + +
Sbjct 695 G--------AEHCSLDDKFRGKTVVDYVTGELSRKPHSVVLLENVEKAEFPDQMRLSEAV 746
Query 122 DEGRLTDSQGRTVDFSNCLLILTSNIG 148
G++ D GR + N ++++TS I
Sbjct 747 STGKIRDLHGRVISMKNVIVVVTSGIA 773
> At2g40130
Length=907
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 57/117 (48%), Gaps = 3/117 (2%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGGQL 89
+GP VGK + +AE ++ ++ R + +D+ + P G +
Sbjct 575 LVGPDTVGKRRMSLVLAEIVYQSEHRFMAVDLGAAEQGMGGC---DDPMRLRGKTMVDHI 631
Query 90 TEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLILTSN 146
E + RNP+ VV L+ +EKA ++ L + ++ G+ DS GR V N + ++TS+
Sbjct 632 FEVMCRNPFCVVFLENIEKADEKLQMSLSKAIETGKFMDSHGREVGIGNTIFVMTSS 688
> YBL022c
Length=1133
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 59/127 (46%), Gaps = 16/127 (12%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVAR-LIGAPPGYVGHEEGGQ 88
F+GP GVGKT + K++A L R M++ E R IGA PG V
Sbjct 630 FVGPPGVGKTSIGKSIARALNRKFFRFSVGGMTDVAEIKGHRRTYIGALPGRVVQ----A 685
Query 89 LTEAVRRNPYSVVLLDEVEK-----AHHQVWNVLLQVLDEGR----LTDSQGRTVDFSNC 139
L + +NP ++L+DE++K H LL+VLD + L + +D S
Sbjct 686 LKKCQTQNP--LILIDEIDKIGHGGIHGDPSAALLEVLDPEQNNSFLDNYLDIPIDLSKV 743
Query 140 LLILTSN 146
L + T+N
Sbjct 744 LFVCTAN 750
> SPCC737.08
Length=4717
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 62/125 (49%), Gaps = 20/125 (16%)
Query 7 VSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYME 66
V +++L A + R PI L GPT GKT + + VA++ T + VR++ E+ +
Sbjct 879 VQKNLLNIARACSTRMFPI---LIQGPTSSGKTSMIEYVAKK---TGHKFVRINNHEHTD 932
Query 67 SHSVARLIGAPPGYVGHEEG------GQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
+ IG YV + G G L EA+ RN Y +V LDE+ A V L ++
Sbjct 933 ---LQEYIGT---YVTDDNGSLSFREGVLVEAL-RNGYWIV-LDELNLAPTDVLEALNRL 984
Query 121 LDEGR 125
LD+ R
Sbjct 985 LDDNR 989
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 66/130 (50%), Gaps = 10/130 (7%)
Query 6 AVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
+ E++ R L ++ +PI L GP G+GK L +A +L ++I+R+ +S+
Sbjct 137 STQENLNRITPYLVQK-RPI---LLAGPEGIGKKFLITQIAAKL---GQQIIRIHLSDST 189
Query 66 ESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGR 125
+ + +P + G LT+AV + +L +E A +V +VLL +L++ +
Sbjct 190 DPKMLIGTYTSPKPGEFEWQPGVLTQAVITGKW--ILFTNIEHAPSEVLSVLLPLLEKRQ 247
Query 126 LT-DSQGRTV 134
L S+G T+
Sbjct 248 LVIPSRGETI 257
> CE16894
Length=971
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 59/129 (45%), Gaps = 20/129 (15%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG-- 87
F GP GVGKT + K++A L R M++ VA + G YVG G
Sbjct 510 FHGPPGVGKTSIAKSIATALNREYFRFSVGGMTD------VAEIKGHRRTYVGAMPGKMI 563
Query 88 QLTEAVR-RNPYSVVLLDEVEKA-----HHQVWNVLLQVLDEGRLTDSQGR----TVDFS 137
Q + V+ NP +VL+DEV+K H + LL++LD + + VD S
Sbjct 564 QCMKKVKTENP--LVLIDEVDKIGGAGFHGDPASALLELLDPEQNANFNDHFLDVPVDLS 621
Query 138 NCLLILTSN 146
L I T+N
Sbjct 622 RVLFICTAN 630
> ECU10g0130
Length=387
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 55/117 (47%), Gaps = 33/117 (28%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G L+ GP G GKT L KAVA TK +R++ SE+++ Y+
Sbjct 166 DPPQGVLLY-GPPGTGKTMLVKAVANH---TKATFIRVNGSEFVQK------------YL 209
Query 82 GHEEGGQLTEAV----RRNPYSVVLLDEVE-----------KAHHQVWNVLLQVLDE 123
G EG ++ V R S+V +DEV+ A +V VL+++L++
Sbjct 210 G--EGPRMVRDVFRLAREKAPSIVFIDEVDSIATKRFDASTSADREVQRVLIELLNQ 264
> ECU08g1970
Length=387
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 55/117 (47%), Gaps = 33/117 (28%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G L+ GP G GKT L KAVA TK +R++ SE+++ Y+
Sbjct 166 DPPQGVLLY-GPPGTGKTMLVKAVANH---TKATFIRVNGSEFVQK------------YL 209
Query 82 GHEEGGQLTEAV----RRNPYSVVLLDEVE-----------KAHHQVWNVLLQVLDE 123
G EG ++ V R S+V +DEV+ A +V VL+++L++
Sbjct 210 G--EGPRMVRDVFRLAREKAPSIVFIDEVDSIATKRFDASTSADREVQRVLIELLNQ 264
> At2g29970
Length=1002
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 63/149 (42%), Gaps = 18/149 (12%)
Query 3 AVTAVSESILRSAAGLARRNKPIGS-----FLFLGPTGVGKTELCKAVAEELFDTKERIV 57
AV A+SE + RRN + + LGP GK ++ A+AE ++ +
Sbjct 645 AVNAISEIVCGYRDESRRRNNHVATTSNVWLALLGPDKAGKKKVALALAEVFCGGQDNFI 704
Query 58 RLDMS--EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWN 115
+D + ++ + + Y+ E V R SVV ++ VEKA
Sbjct 705 CVDFKSQDSLDDRFRGKTVV---DYIAGE--------VARRADSVVFIENVEKAEFPDQI 753
Query 116 VLLQVLDEGRLTDSQGRTVDFSNCLLILT 144
L + + G+L DS GR + N +++ T
Sbjct 754 RLSEAMRTGKLRDSHGREISMKNVIVVAT 782
> ECU08g1900
Length=2832
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 68/149 (45%), Gaps = 12/149 (8%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
+T ++ LR+ N P+ L G T GKT + ++ E+ R++R++
Sbjct 636 VLTPRIQTYLRTMRQAILSNCPL---LLQGDTSTGKTSMILFLSREMC---RRVIRINNH 689
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ E+ + + +EG L A+RR + V+LDE+ A V VL ++LD
Sbjct 690 EHTEAADYIGSFASTSDGIEFKEGA-LVGAMRRGDW--VILDELNLACSDVLEVLNRLLD 746
Query 123 EGR---LTDSQGRTVDFSNCLLILTSNIG 148
+ R + ++ V N + T NIG
Sbjct 747 DNRQIYIPETDEVVVPHENFRIFATQNIG 775
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 46/98 (46%), Gaps = 8/98 (8%)
Query 29 LFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHS-VARLIGAPPGYVGHEEGG 87
L +G TG+GKT+ C +A +F T R++ L M +ES + + V + G
Sbjct 957 LIVGETGIGKTKACD-IASSVFKT--RLITLSMHRGIESSDFIGSFVFENSKIVWKD--G 1011
Query 88 QLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGR 125
L +A++ L+DE+ A V L VL+ R
Sbjct 1012 PLVKAMKCG--DAFLIDEINLAEDSVLERLNSVLESQR 1047
> HsM4758682
Length=937
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 65/151 (43%), Gaps = 16/151 (10%)
Query 7 VSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
V + IL A R G L F GP GVGKT + +++A L R M++
Sbjct 475 VKKRILEFIAVSQLRGSTQGKILCFYGPPGVGKTSIARSIARALNREYFRFSVGGMTDVA 534
Query 66 ESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKA----HHQVWNVLLQV 120
E R +GA PG + L + NP ++L+DEV+K + LL++
Sbjct 535 EIKGHRRTYVGAMPGKIIQ----CLKKTKTENP--LILIDEVDKIGRGYQGDPSSALLEL 588
Query 121 LDEGR----LTDSQGRTVDFSNCLLILTSNI 147
LD + L VD S L I T+N+
Sbjct 589 LDPEQNANFLDHYLDVPVDLSKVLFICTANV 619
> Hs21396489
Length=959
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 65/151 (43%), Gaps = 16/151 (10%)
Query 7 VSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
V + IL A R G L F GP GVGKT + +++A L R M++
Sbjct 497 VKKRILEFIAVSQLRGSTQGKILCFYGPPGVGKTSIARSIARALNREYFRFSVGGMTDVA 556
Query 66 ESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKA----HHQVWNVLLQV 120
E R +GA PG + L + NP ++L+DEV+K + LL++
Sbjct 557 EIKGHRRTYVGAMPGKIIQ----CLKKTKTENP--LILIDEVDKIGRGYQGDPSSALLEL 610
Query 121 LDEGR----LTDSQGRTVDFSNCLLILTSNI 147
LD + L VD S L I T+N+
Sbjct 611 LDPEQNANFLDHYLDVPVDLSKVLFICTANV 641
> At5g47040
Length=888
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 52/101 (51%), Gaps = 17/101 (16%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGGQL 89
F+GP GVGKT L ++A L + VRL + + A + G Y+G G+L
Sbjct 406 FVGPPGVGKTSLASSIAAAL---GRKFVRLSLGGVKDE---ADIRGHRRTYIGSMP-GRL 458
Query 90 TEAVRR----NPYSVVLLDEVEKAHHQV----WNVLLQVLD 122
+ ++R NP V+LLDE++K V + LL+VLD
Sbjct 459 IDGLKRVGVCNP--VMLLDEIDKTGSDVRGDPASALLEVLD 497
> SPAC2G11.06
Length=432
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 17/87 (19%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDM-SEYM-ESHSVARLIGAPPGY 80
KP L GP G GK+ L KAVA E T I D+ S++M ES + R
Sbjct 160 KPWSGILLYGPPGTGKSYLAKAVATEAGSTFFSISSSDLVSKWMGESERLVR-------- 211
Query 81 VGHEEGGQLTEAVRRNPYSVVLLDEVE 107
QL E R S++ +DE++
Sbjct 212 -------QLFEMAREQKPSIIFIDEID 231
> 7293766
Length=1006
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 61/130 (46%), Gaps = 21/130 (16%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGGQL 89
F GP GVGKT + K++A L R M++ VA + G YVG G+L
Sbjct 544 FHGPPGVGKTSIAKSIARALNREYFRFSVGGMTD------VAEIKGHRRTYVG-AMPGKL 596
Query 90 TEAVRR----NPYSVVLLDEVEKA----HHQVWNVLLQVLDEGR----LTDSQGRTVDFS 137
+ +++ NP +VL+DEV+K + LL++LD + L VD S
Sbjct 597 IQCLKKTKIENP--LVLIDEVDKIGKGYQGDPSSALLELLDPEQNANFLDHYLDVPVDLS 654
Query 138 NCLLILTSNI 147
L I T+N+
Sbjct 655 RVLFICTANV 664
> YDR394w
Length=428
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 22/90 (24%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G L+ GP G GKT L KAVA TK +R++ SE++ Y+
Sbjct 204 DPPRGVLLY-GPPGTGKTMLVKAVANS---TKAAFIRVNGSEFVHK------------YL 247
Query 82 GHEEGGQLTEAV----RRNPYSVVLLDEVE 107
G EG ++ V R N S++ +DEV+
Sbjct 248 G--EGPRMVRDVFRLARENAPSIIFIDEVD 275
> YPR173c
Length=437
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 39/87 (44%), Gaps = 17/87 (19%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDM-SEYM-ESHSVARLIGAPPGY 80
KP L GP G GK+ L KAVA E T + D+ S++M ES + +
Sbjct 164 KPTSGILLYGPPGTGKSYLAKAVATEANSTFFSVSSSDLVSKWMGESEKLVK-------- 215
Query 81 VGHEEGGQLTEAVRRNPYSVVLLDEVE 107
QL R N S++ +DEV+
Sbjct 216 -------QLFAMARENKPSIIFIDEVD 235
> CE23016
Length=773
Score = 37.0 bits (84), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 55/117 (47%), Gaps = 22/117 (18%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERI----V 57
A+ V E +L A N G L F GP G+GKT + KA+AE + +R+ +
Sbjct 306 AMDDVKERVLEHLAVCKMNNSVKGMILCFTGPPGIGKTSIAKAIAESMGRKFQRVSLGGI 365
Query 58 RLDMSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVR----RNPYSVVLLDEVEKAH 110
R D S+ + H + P G++ EA++ NP V LLDEV+K +
Sbjct 366 R-DESD-IRGHRRTYVAAMP---------GRIIEALKTCKTNNP--VFLLDEVDKLY 409
> At3g05790
Length=942
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 46/155 (29%), Positives = 69/155 (44%), Gaps = 17/155 (10%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFL-GPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
++ V E IL A R G + L GPTGVGKT + +++A L R +S
Sbjct 427 LSDVKERILEFIAVGGLRGTSQGKIICLSGPTGVGKTSIGRSIARALDRKFFRFSVGGLS 486
Query 63 EYMESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEK----AHH-QVWNV 116
+ E R IGA PG + L NP +VL+DE++K HH +
Sbjct 487 DVAEIKGHRRTYIGAMPGKMVQ----CLKNVGTENP--LVLIDEIDKLGVRGHHGDPASA 540
Query 117 LLQVLDEGR----LTDSQGRTVDFSNCLLILTSNI 147
+L++LD + L +D S L + T+N+
Sbjct 541 MLELLDPEQNANFLDHYLDVPIDLSKVLFVCTANV 575
> 7299244
Length=421
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 52/113 (46%), Gaps = 25/113 (22%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G LF GP G GKT L KAVA T +R+ SE+ V + +G P V
Sbjct 200 DPPRGVLLF-GPPGCGKTMLAKAVAHH---TTASFIRVVGSEF-----VQKYLGEGPRMV 250
Query 82 GHEEGGQLTEAVRRNPYSVVLLDEVEK-----------AHHQVWNVLLQVLDE 123
L ++N S++ +DE++ A +V +LL++L++
Sbjct 251 -----RDLFRLAKQNSPSIIFIDEIDAIATKRFDAQTGADREVQRILLELLNQ 298
> At5g40090
Length=459
Score = 36.2 bits (82), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 44/95 (46%), Gaps = 2/95 (2%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
NK + + G GVGKT L + + E+F + V LD E M+ + P +
Sbjct 200 NKEVRTIGIWGSAGVGKTTLARYIYAEIFVNFQTHVFLDNVENMKDKLLKFEGEEDPTVI 259
Query 82 --GHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVW 114
+ +G ++TEA R++ +++ D+V W
Sbjct 260 ISSYHDGHEITEARRKHRKILLIADDVNNMEQGKW 294
> At1g62130
Length=372
Score = 36.2 bits (82), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 41/95 (43%), Gaps = 27/95 (28%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVG 82
+P L GP+G GKT L KAVA E ++ + MS +
Sbjct 135 QPCNGILLFGPSGTGKTMLAKAVATE---AGANLINMSMSRWF----------------- 174
Query 83 HEEGGQLTEAV-----RRNPYSVVLLDEVEKAHHQ 112
EG + +AV + +P S++ LDEVE H+
Sbjct 175 -SEGEKYVKAVFSLASKISP-SIIFLDEVESMLHR 207
> At2g27600
Length=435
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 41/87 (47%), Gaps = 13/87 (14%)
Query 21 RNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGY 80
+ +P +FL GP G GK+ L KAVA E T + D+ V++ +G
Sbjct 161 KRRPWRAFLLYGPPGTGKSYLAKAVATEADSTFFSVSSSDL--------VSKWMGESEKL 212
Query 81 VGHEEGGQLTEAVRRNPYSVVLLDEVE 107
V + L E R + S++ +DE++
Sbjct 213 VSN-----LFEMARESAPSIIFVDEID 234
> YLR106c
Length=4910
Score = 35.0 bits (79), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 52/103 (50%), Gaps = 17/103 (16%)
Query 29 LFLGPTGVGKTELCKAVAEELFDTKERIVRL------DMSEYMESHSVARLIGAPPGYVG 82
L GPT GKT + K +A+ T + VR+ D+ EY+ ++ + G +
Sbjct 1080 LIQGPTSSGKTSMIKYLADI---TGHKFVRINNHEHTDLQEYLGTY-----VTDDTGKLS 1131
Query 83 HEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGR 125
+EG L EA+R+ + ++LDE+ A V L ++LD+ R
Sbjct 1132 FKEGV-LVEALRKGYW--IVLDELNLAPTDVLEALNRLLDDNR 1171
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 68/135 (50%), Gaps = 15/135 (11%)
Query 18 LARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLI--- 74
L R+ + + +G G GKT L +++ + + IV++ + E + A+L+
Sbjct 301 LGRKIQNSTPIMLIGKAGSGKTFLINELSKYM-GCHDSIVKIHLGE----QTDAKLLIGT 355
Query 75 ---GAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLT-DSQ 130
G PG G L AV+ + VL+++++KA V ++LL +L++ LT S+
Sbjct 356 YTSGDKPG-TFEWRAGVLATAVKEGRW--VLIEDIDKAPTDVLSILLSLLEKRELTIPSR 412
Query 131 GRTVDFSNCLLILTS 145
G TV +N ++++
Sbjct 413 GETVKAANGFQLIST 427
> Hs20562018
Length=852
Score = 35.0 bits (79), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 48/154 (31%), Positives = 70/154 (45%), Gaps = 16/154 (10%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDM 61
A+ + + +L A +N G L F+GP GVGKT + ++VA+ L RI +
Sbjct 345 AMEKLKKRVLEYLAVRQLKNNLKGPILCFVGPPGVGKTSVGRSVAKTLGREFHRIALGGV 404
Query 62 SEYMESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWN----V 116
+ + R +G+ PG + + G T V NP V LLDEV+K +
Sbjct 405 CDQSDIRGHRRTYVGSMPGRIIN---GLKTVGV-NNP--VFLLDEVDKLGKSLQGDPAAA 458
Query 117 LLQVLDEGR---LTDSQ-GRTVDFSNCLLILTSN 146
LL+VLD + TD D S L I T+N
Sbjct 459 LLEVLDPEQNHNFTDHYLNVAFDLSQVLFIATAN 492
> At3g13470
Length=596
Score = 34.7 bits (78), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 44/89 (49%), Gaps = 7/89 (7%)
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRR-NPYSVVLLDEVEKAHHQVWNVLLQ 119
MS+ +E +A + G HE G + EA+ + VV L+E + A + ++ V
Sbjct 185 MSKEVEDSELADVAAVSAGN-NHEVGSMIAEAMSKVGRKGVVTLEEGKSAENNLYVVEGM 243
Query 120 VLDEGRL-----TDSQGRTVDFSNCLLIL 143
D G + TDS+ +V++ NC L+L
Sbjct 244 QFDRGYISPYFVTDSEKMSVEYDNCKLLL 272
> YJR068w
Length=353
Score = 34.3 bits (77), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 49/114 (42%), Gaps = 19/114 (16%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFD---TKERIVRLD 60
VTA ++ L N P LF GP G GKT A+ +EL+ K RI+ L+
Sbjct 39 VTAQDHAVTVLKKTLKSANLP--HMLFYGPPGTGKTSTILALTKELYGPDLMKSRILELN 96
Query 61 MSEYM-------ESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVE 107
S+ + + ARL + P H+ E PY +++LDE +
Sbjct 97 ASDERGISIVREKVKNFARLTVSKPS--KHD-----LENYPCPPYKIIILDEAD 143
> CE20667
Length=1302
Score = 34.3 bits (77), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 57/115 (49%), Gaps = 7/115 (6%)
Query 21 RNKPIGSFLFL-GPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPG 79
R+ +GS L L G GVGK +L L + + ++L +++ ++ ++ G
Sbjct 661 RDMKLGSHLLLIGNQGVGKNKLTDRFLH-LINRPRQYMQLHRDTTVQTLTMQTVVEN--G 717
Query 80 YVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTV 134
+ HE+ L +A R ++++DE +KA V +L +LD G L GRT+
Sbjct 718 IIRHEDSA-LVKAARSG--QILVIDEADKAPLHVIAILKSLLDTGTLVLGDGRTL 769
> CE01547
Length=424
Score = 34.3 bits (77), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 9/85 (10%)
Query 28 FLFLGPTGVGKTELCKAVAEELF----DTKERIVRLDMSEYMESHSVARLIGAPPGYVGH 83
L GP G GKT LCK +A+ L D + V L+++ SHS+ + G +
Sbjct 175 ILLTGPPGTGKTSLCKGLAQHLSIRMNDKYSKSVMLEIN----SHSLFSKWFSESGKLVQ 230
Query 84 EEGGQLTEAVRRNPYSV-VLLDEVE 107
+ Q+ E V VL+DEVE
Sbjct 231 KMFDQIDELAEDEKCMVFVLIDEVE 255
> At4g24850
Length=442
Score = 34.3 bits (77), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 38/85 (44%), Gaps = 13/85 (15%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVG 82
KP L GP G GKT L KAVA+E + + MS ++ G YV
Sbjct 173 KPCKGILLFGPPGTGKTMLAKAVAKE---ADANFINISMSSI-----TSKWFGEGEKYV- 223
Query 83 HEEGGQLTEAVRRNPYSVVLLDEVE 107
+ A + +P SV+ +DEV+
Sbjct 224 ---KAVFSLASKMSP-SVIFVDEVD 244
> CE20666
Length=1767
Score = 34.3 bits (77), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 57/115 (49%), Gaps = 7/115 (6%)
Query 21 RNKPIGSFLFL-GPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPG 79
R+ +GS L L G GVGK +L L + + ++L +++ ++ ++ G
Sbjct 661 RDMKLGSHLLLIGNQGVGKNKLTDRFLH-LINRPRQYMQLHRDTTVQTLTMQTVVEN--G 717
Query 80 YVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTV 134
+ HE+ L +A R ++++DE +KA V +L +LD G L GRT+
Sbjct 718 IIRHEDSA-LVKAARSG--QILVIDEADKAPLHVIAILKSLLDTGTLVLGDGRTL 769
> At5g58290
Length=408
Score = 33.9 bits (76), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 38/140 (27%), Positives = 64/140 (45%), Gaps = 39/140 (27%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G L+ GP G GKT L KAVA T +R+ SE+++ Y+
Sbjct 187 DPPRGVLLY-GPPGTGKTMLAKAVANH---TTAAFIRVVGSEFVQK------------YL 230
Query 82 GHEEGGQLTEAVRR----NPYSVVLLDEVEK-----------AHHQVWNVLLQVLDEGRL 126
G EG ++ V R N +++ +DEV+ A +V +L+++L++
Sbjct 231 G--EGPRMVRDVFRLAKENAPAIIFIDEVDAIATARFDAQTGADREVQRILMELLNQ--- 285
Query 127 TDSQGRTVDFSNCLLILTSN 146
D +TV N +I+ +N
Sbjct 286 MDGFDQTV---NVKVIMATN 302
> 7303557
Length=4865
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 41/146 (28%), Positives = 66/146 (45%), Gaps = 13/146 (8%)
Query 7 VSESILRSAAGLARRNKPIGSF--LFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEY 64
++ES+ ++ LAR IG L GPT GKT L VA + R +R++ E+
Sbjct 1090 LTESVKKNLKDLARI-ISIGKLPILLQGPTSAGKTSLIDYVARR---SGNRCLRINNHEH 1145
Query 65 MESHSVARLIGAP-PGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDE 123
+ A G + EG L +A+R + ++LDE+ A + L +VLD+
Sbjct 1146 TDLQEYIGTYAADLDGKLTFREGV-LVQAMRHGFW--IILDELNLASTDILEALNRVLDD 1202
Query 124 GR---LTDSQGRTVDFSNCLLILTSN 146
R + ++Q N +L T N
Sbjct 1203 NRELYIAETQTLVKAHPNFMLFATQN 1228
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 29 LFLGPTGVGKTELCKAVAEELFDTKERIVRLDM 61
L +GPTG GKT +C+ +A + D + RI+ M
Sbjct 1420 LLVGPTGCGKTTVCQLLA-SIADVQLRILNCHM 1451
> At1g73170
Length=652
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 12/89 (13%)
Query 27 SFLFLGPTGVGKTELCKAVAEEL-FDTKERIVRLDMSEYMESH--------SVARLIGAP 77
S L +GP GVGKT + + VA L D ++R++ +D S + AR + P
Sbjct 199 SLLLIGPPGVGKTTMIREVARMLGNDYEKRVMIVDTSNEIGGDGDIPHPGIGNARRMQVP 258
Query 78 PGYVGHEEGGQLTEAVRRNPYSVVLLDEV 106
+ H+ L EAV + V+++DE+
Sbjct 259 NSDIQHK---VLIEAVENHMPQVIVIDEI 284
> At3g05780
Length=924
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 70/157 (44%), Gaps = 22/157 (14%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFL-GPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
++ V E IL A R G + L GP GVGKT + +++A L + R +
Sbjct 418 LSDVKERILEFIAVGRLRGTSQGKIICLSGPPGVGKTSIGRSIARAL---DRKFFRFSVG 474
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRR----NPYSVVLLDEVEKAHH----QVW 114
+ VA + G YVG G++ + ++ NP ++L DE++K
Sbjct 475 GLSD---VAEIKGHCQTYVG-AMPGKMVQCLKSVGTANP--LILFDEIDKLGRCHTGDPA 528
Query 115 NVLLQVLDEGR----LTDSQGRTVDFSNCLLILTSNI 147
+ LL+V+D + L T+D S L + T+N+
Sbjct 529 SALLEVMDPEQNAKFLDHFLNVTIDLSKVLFVCTANV 565
Lambda K H
0.318 0.135 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40