bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5454_orf3
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
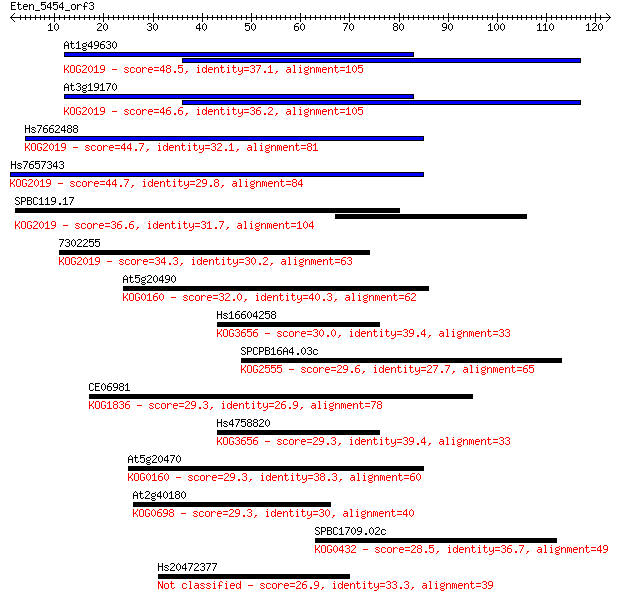
At1g49630 48.5 3e-06
At3g19170 46.6 1e-05
Hs7662488 44.7 4e-05
Hs7657343 44.7 5e-05
SPBC119.17 36.6 0.011
7302255 34.3 0.063
At5g20490 32.0 0.29
Hs16604258 30.0 1.2
SPCPB16A4.03c 29.6 1.3
CE06981 29.3 1.7
Hs4758820 29.3 1.8
At5g20470 29.3 1.8
At2g40180 29.3 1.9
SPBC1709.02c 28.5 3.4
Hs20472377 26.9 8.2
> At1g49630
Length=1076
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 44/73 (60%), Gaps = 2/73 (2%)
Query 12 YNRDPLGALEFEQDIWRLRERLA--AGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQ 69
Y+ DP L++E+ + L+ R+A + VF +++E +L NPH VT+ M+ E A +
Sbjct 512 YDMDPFEPLKYEEPLKSLKARIAEKGSKSVFSPLIEEYILNNPHCVTIEMQPDPEKASLE 571
Query 70 AAVERLVLQTLEA 82
A E+ +L+ ++A
Sbjct 572 EAEEKSILEKVKA 584
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 4/81 (4%)
Query 36 GEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQAAVERLVLQTLEAIAAKGFSAAEIEA 95
GE + + +++ LL+ + L+ S + VE LV+ TL +A +GF +EA
Sbjct 424 GEALVNSGMEDELLQPQFSIGLKGVSD----DNVQKVEELVMNTLRKLADEGFDTDAVEA 479
Query 96 AFNKVDFSLRETPRKELPRGL 116
+ N ++FSLRE PRGL
Sbjct 480 SMNTIEFSLRENNTGSSPRGL 500
> At3g19170
Length=1052
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 44/73 (60%), Gaps = 2/73 (2%)
Query 12 YNRDPLGALEFEQDIWRLRERLA--AGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQ 69
Y+ DP L++ + + L+ R+A + VF ++++L+L N HRVT+ M+ E A Q+
Sbjct 489 YDMDPFEPLKYTEPLKALKTRIAEEGSKAVFSPLIEKLILNNSHRVTIEMQPDPEKATQE 548
Query 70 AAVERLVLQTLEA 82
E+ +L+ ++A
Sbjct 549 EVEEKNILEKVKA 561
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 4/81 (4%)
Query 36 GEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQAAVERLVLQTLEAIAAKGFSAAEIEA 95
GE + + + + LL+ + L+ S E VE L++ TL+ +A +GF +EA
Sbjct 401 GEALVSSGLSDELLQPQFGIGLKGVSE----ENVQKVEELIMDTLKKLAEEGFDNDAVEA 456
Query 96 AFNKVDFSLRETPRKELPRGL 116
+ N ++FSLRE PRGL
Sbjct 457 SMNTIEFSLRENNTGSFPRGL 477
> Hs7662488
Length=595
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 44/82 (53%), Gaps = 1/82 (1%)
Query 4 QRMAQELNYNRDPLGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSA 63
Q +A N++ DP+ L+ + + R+ L K + VK+ N H++TL M+
Sbjct 7 QYIASCWNHDGDPVELLKLGNQLAKFRQCLQENPKFLQEKVKQYFKNNQHKLTLSMRPDD 66
Query 64 EYAEQQAAVERLVL-QTLEAIA 84
+Y E+QA VE L Q +EA++
Sbjct 67 KYHEKQAQVEATKLKQKVEALS 88
> Hs7657343
Length=1038
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 1 IFSQRMAQELNYNRDPLGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMK 60
+ + +A N++ DP+ L+ + + R+ L K + VK+ N H++TL M+
Sbjct 446 MLTSYIASCWNHDGDPVELLKLGNQLAKFRQCLQENPKFLQEKVKQYFKNNQHKLTLSMR 505
Query 61 SSAEYAEQQAAVERLVL-QTLEAIA 84
+Y E+QA VE L Q +EA++
Sbjct 506 PDDKYHEKQAQVEATKLKQKVEALS 530
> SPBC119.17
Length=882
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 45/84 (53%), Gaps = 7/84 (8%)
Query 2 FSQRMAQELNYN----RDPLGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVTL 57
F +AQ L +N DP L F + I L+++ + G K+F+ ++K+ +LEN R
Sbjct 428 FGIGLAQSLPFNWFNGADPADWLSFNKQIEWLKQKNSDG-KLFQKLIKKYILENKSRFVF 486
Query 58 RMKSSAEYAE--QQAAVERLVLQT 79
M S+ + + Q+A ++L +T
Sbjct 487 TMLPSSTFPQRLQEAEAKKLQERT 510
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 67 EQQAAVERLVLQTLEAIAAKGFSAAEIEAAFNKVDFSLR 105
E A +E LV +A KGF ++EA ++++ SL+
Sbjct 384 ESLAKIENLVYSIFNDLALKGFENEKLEAILHQMEISLK 422
> 7302255
Length=1112
Score = 34.3 bits (77), Expect = 0.063, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 11 NYNRDPLGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEYAE--Q 68
N++ D + L I LRE ++ +K F+ +++ N HR+TL M Y + +
Sbjct 481 NHDGDVVSNLRVSDMISGLRESISQNKKYFQEKIEKYFANNNHRLTLTMSPDEAYEDKFK 540
Query 69 QAAVE 73
QA +E
Sbjct 541 QAELE 545
> At5g20490
Length=1544
Score = 32.0 bits (71), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 24 QDIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQAAVERLVLQTLEAI 83
+D LRE EK E + L LE R L + EYA+QQ A+E + LQ EA
Sbjct 900 RDTGALREAKDKLEKRVEELTWRLQLEKRQRTELEEAKTQEYAKQQEALETMRLQVEEAN 959
Query 84 AA 85
AA
Sbjct 960 AA 961
> Hs16604258
Length=420
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 43 MVKELLLENPHRVTLRMKSSAEYAEQQAAVERL 75
+V+E +NPH TL + SAE +Q+ +E L
Sbjct 379 LVQESTFQNPHGETLLYRKSAEKPQQELVMEEL 411
> SPCPB16A4.03c
Length=585
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 34/65 (52%), Gaps = 3/65 (4%)
Query 48 LLENPHRVTLRMKSSAEYAEQQAAVERLVLQTLEAIAAKGFSAAEIEAAFNKVDFSLRET 107
L +P + ++ K SA+ E+ A++ L L+A+ A+G + E+AF + L +
Sbjct 459 LRHHPKVLGMQFKKSAKRPEKSNAID---LYVLDAVPAEGSEREQWESAFETIPEPLTKK 515
Query 108 PRKEL 112
R+E
Sbjct 516 EREEF 520
> CE06981
Length=1557
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 32/78 (41%), Gaps = 0/78 (0%)
Query 17 LGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQAAVERLV 76
+GA E D+ R + + EN++ L E R + +S +Y EQ + L
Sbjct 1125 VGADAAENDMKRWEIIIENARREIENVLHYLETEGEERAQIAYNASQKYGEQSKRMSELA 1184
Query 77 LQTLEAIAAKGFSAAEIE 94
T E A+EIE
Sbjct 1185 SGTREEAEKHLKQASEIE 1202
> Hs4758820
Length=522
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 43 MVKELLLENPHRVTLRMKSSAEYAEQQAAVERL 75
+V+E +NPH TL + SAE +Q+ +E L
Sbjct 481 LVQESTFQNPHGETLLYRKSAEKPQQELVMEEL 513
> At5g20470
Length=556
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 25 DIWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQAAVERLVLQTLEAIA 84
D LRE EK E + L LE R L + EYA+QQ A++ + LQ EA A
Sbjct 102 DTGALREAKDKLEKRVEELTLRLQLETRQRTDLEEAKTQEYAKQQEALQAMWLQVEEANA 161
> At2g40180
Length=390
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 26 IWRLRERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEY 65
+WR++ LA + + +KE ++ P TLR+K E+
Sbjct 286 VWRIQGTLAVSRGIGDRYLKEWVIAEPETRTLRIKPEFEF 325
> SPBC1709.02c
Length=980
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 9/58 (15%)
Query 63 AEYAEQQAAVERLVLQTLEAIAAKGFSAAEIEAA---------FNKVDFSLRETPRKE 111
AEY E+ E+ VLQ L++ A K ++ +E+A F + +F P+KE
Sbjct 72 AEYVEKTTPGEKKVLQDLDSPALKSYNPKAVESAWYDWWVKSGFFEPEFGPDGKPKKE 129
> Hs20472377
Length=465
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 31 ERLAAGEKVFENMVKELLLENPHRVTLRMKSSAEYAEQQ 69
++ AAG +FEN + + LE P KS+ +QQ
Sbjct 108 QKYAAGSSIFENFIYQSSLEIPFGSICEAKSATAILQQQ 146
Lambda K H
0.319 0.132 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40