bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5447_orf1
Length=187
Score E
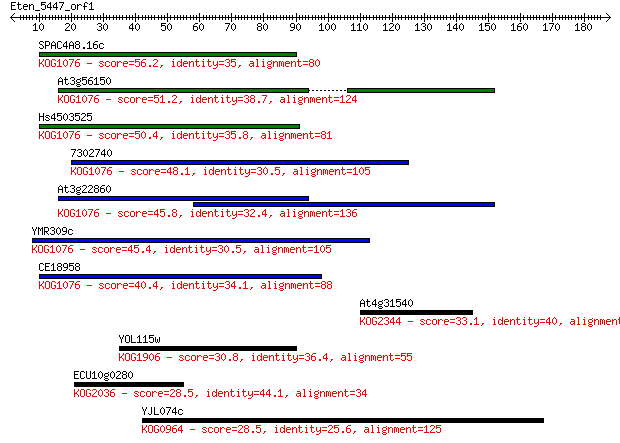
Sequences producing significant alignments: (Bits) Value
SPAC4A8.16c 56.2 4e-08
At3g56150 51.2 1e-06
Hs4503525 50.4 2e-06
7302740 48.1 9e-06
At3g22860 45.8 5e-05
YMR309c 45.4 7e-05
CE18958 40.4 0.002
At4g31540 33.1 0.32
YOL115w 30.8 1.9
ECU10g0280 28.5 8.8
YJL074c 28.5 9.1
> SPAC4A8.16c
Length=639
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
Q+ S++ SF+ERLDD+ T+SLQ D H+ EY +RL + LL R+ +L+ G
Sbjct 167 QVQGSVV-SFLERLDDEFTRSLQMIDPHTPEYIDRLKDETSLYTLLVRSQGYLERIGVVE 225
Query 70 QAATVALKVNEHMHYKPDTI 89
A + ++ + ++YKP+ +
Sbjct 226 NTARLIMRRLDRVYYKPEQV 245
> At3g56150
Length=900
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 16 LTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERG-FKAQAATV 74
L +F+ER+D + KSLQ D H+ EY ERL LAL + + G FKA AA V
Sbjct 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKA-AAKV 414
Query 75 ALKVNEHMHYKPDTIASKM 93
AL+ E ++YKP + M
Sbjct 415 ALRRVEAIYYKPQEVYDAM 433
Score = 36.6 bits (83), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 106 SPQCHLEVLGHLISAEFDTTSGVFTSMPLQIWMETFAGVKLLLSIM 151
+P LE+L +ISA+FD G+ MP+ +W + + +L I+
Sbjct 276 TPAQKLEILFSVISAQFDVNPGLSGHMPINVWKKCVLNMLTILDIL 321
> Hs4503525
Length=913
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKA 69
++ ILT VER+D++ TK +Q+TD HS EY E L + A++ R +L+E+G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 497
Query 70 QAATVALKVNEHMHYKPDTIA 90
+ + L H +YK D A
Sbjct 498 EVCRIYLLRILHTYYKFDYKA 518
> 7302740
Length=910
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 53/108 (49%), Gaps = 8/108 (7%)
Query 20 VERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKAQAATVALKVN 79
VERLDD+ K L+ D HS++Y RL ++++ + + + G + + L+
Sbjct 425 VERLDDEFVKLLKECDPHSNDYVSRLKDEVNVVKTIELVLQYFERSGTNNERCRIYLRKI 484
Query 80 EHMHYK--PDTIASKMWDVLRKELAETVSPQCH-LEVLGHLISAEFDT 124
EH++YK P+ + K R EL T S ++ L I A+ DT
Sbjct 485 EHLYYKFDPEVLKKK-----RGELPATTSTSVDVMDKLCKFIYAKDDT 527
> At3g22860
Length=800
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 16 LTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKAQAATVA 75
L +F+E+++ + KSLQ D H+++Y ERL LAL +L+ G A+ VA
Sbjct 313 LVAFLEKIETEFFKSLQCIDPHTNDYVERLKDEPMFLALAQSIQDYLERTGDSKAASKVA 372
Query 76 LKVNEHMHYKPDTIASKM 93
+ E ++YKP + M
Sbjct 373 FILVESIYYKPQEVFDAM 390
Score = 35.0 bits (79), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 53/96 (55%), Gaps = 6/96 (6%)
Query 58 TFCFLDERGFKAQAATVALKVNEHMHYKP-DTIASKMWDVLRKELAETVSPQCHLEVLGH 116
T+ ++++ + +AA + + + + KP +T A K D+ + +A+T P +E+L
Sbjct 187 TWNMVNKKFKEIRAARWSKRRSSSLKLKPGETHAQKHMDLTK--IAKT--PAQKVEILFS 242
Query 117 LISAEFDTTSGVFTS-MPLQIWMETFAGVKLLLSIM 151
+ISAEF+ SG + MP+ +W + + +L I+
Sbjct 243 VISAEFNVNSGGLSGYMPIDVWKKCVVNMLTILDIL 278
> YMR309c
Length=812
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 55/114 (48%), Gaps = 10/114 (8%)
Query 8 IQQMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFL----- 62
++++ SI SFVERLDD+ KSL + D HS +Y RL I L+ RT +
Sbjct 362 VKRILGSIF-SFVERLDDEFMKSLLNIDPHSSDYLIRLRDEQSIYNLILRTQLYFEATLK 420
Query 63 DERGFKAQAATVALKVNEHMHYKPDTIASKM----WDVLRKELAETVSPQCHLE 112
DE + +K +H++YK + + M W+++ + + + L+
Sbjct 421 DEHDLERALTRPFVKRLDHIYYKSENLIKIMETAAWNIIPAQFKSKFTSKDQLD 474
> CE18958
Length=898
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 51/97 (52%), Gaps = 10/97 (10%)
Query 10 QMSASILTSFVERLDDDLTKSLQSTDVHSDEYKERLGKSIDILALLWRTFCFLDERG--- 66
++ SIL + V+RLD +L K LQ+ D HS++Y E+L D+ +L+ + +++ R
Sbjct 414 RIQGSILIA-VQRLDGELAKILQNADCHSNDYIEKLKAEKDMCSLIEKAEKYVELRNDSG 472
Query 67 --FKAQAATVALKVNEHMHYK----PDTIASKMWDVL 97
K + V + EH +YK + A K+ D L
Sbjct 473 IFDKHEVCKVYMMRIEHAYYKYQDQNEEDAGKLMDYL 509
> At4g31540
Length=686
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 110 HLE-VLGHLISAEFDTTSGVFTSMPLQIWMETFAGV 144
HLE + HL AEF + VF + L +WM+ F+ +
Sbjct 298 HLEFAVKHLFEAEFKLCNDVFERLGLNVWMDCFSKI 333
> YOL115w
Length=584
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 35 DVHSDEYKERLGKSI--DILALLWRTFCFLDERGFKAQAATVALKVNEHMHYKPDTI 89
++HS +K+RLGKSI +++ + F DERG A + NE+ H K I
Sbjct 460 ELHSATFKDRLGKSILGNVIKYRGKARDFKDERGLVLNKAIIE---NENYHKKRSRI 513
> ECU10g0280
Length=825
Score = 28.5 bits (62), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 21 ERLDDDLTKSLQSTDVHSDEYKER---LGKSIDILAL 54
E +DDL+ ++S D+ EYKE LG+++D+L L
Sbjct 82 EESEDDLSLFIKSNDIEFIEYKESERILGRTVDMLIL 118
> YJL074c
Length=1230
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 32/134 (23%), Positives = 64/134 (47%), Gaps = 14/134 (10%)
Query 42 KERLGKSIDILALLWRTFCFLDERGFKAQAATVALKVNEHMHYKPDTIASKMWD--VLRK 99
K++L +S+D LWR E+ + T+ VN++ +T++ + + + K
Sbjct 469 KQKLSESLDTRKELWRK-----EQKLQTVLETLLSDVNQNQRNVNETMSRSLANGIINVK 523
Query 100 ELAE--TVSPQCHLEVLGHLI--SAEFDTTSGVFTSMPL-QIWMETFAGVKLLLSIMEQN 154
E+ E +SP+ LG LI + ++ T + V L I ++T L+++ + +
Sbjct 524 EITEKLKISPESVFGTLGELIKVNDKYKTCAEVIGGNSLFHIVVDTEETATLIMNELYRM 583
Query 155 KGAYL--VPLAGLA 166
KG + +PL L+
Sbjct 584 KGGRVTFIPLNRLS 597
Lambda K H
0.316 0.129 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40