bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5430_orf4
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
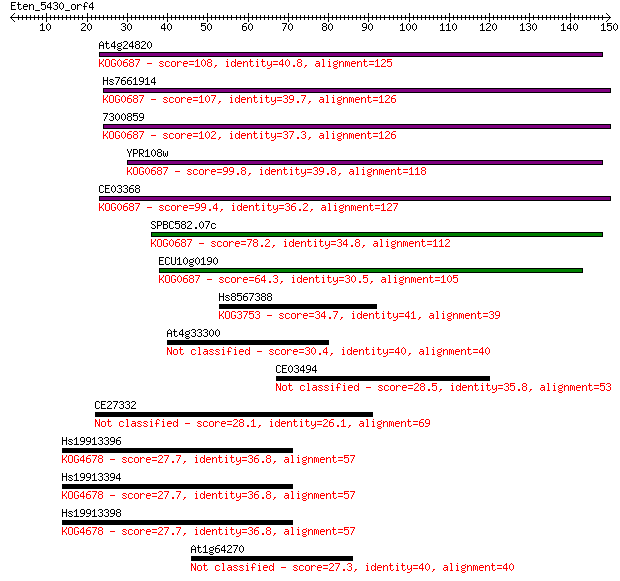
At4g24820 108 5e-24
Hs7661914 107 6e-24
7300859 102 3e-22
YPR108w 99.8 2e-21
CE03368 99.4 2e-21
SPBC582.07c 78.2 5e-15
ECU10g0190 64.3 8e-11
Hs8567388 34.7 0.063
At4g33300 30.4 1.2
CE03494 28.5 4.3
CE27332 28.1 6.5
Hs19913396 27.7 8.8
Hs19913394 27.7 9.1
Hs19913398 27.7 9.4
At1g64270 27.3 9.9
> At4g24820
Length=406
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 51/125 (40%), Positives = 81/125 (64%), Gaps = 0/125 (0%)
Query 23 EAYRVAYAKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLK 82
E ++ KTV +G ++D++ L+ Y D DLV K + AK+ E+GGDWER+N+LK
Sbjct 120 EQLKLTEGKTVAVGQKMDVVFYTLQLAFFYMDFDLVSKSIDKAKKLFEEGGDWERKNRLK 179
Query 83 VIEALDFILSRNFTEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVV 142
V E L + +RNF +A+ L L+ ++TF + I +ETF+FY L +++ DR +LK++VV
Sbjct 180 VYEGLYCMSTRNFKKAASLFLDSISTFTTYEIFPYETFIFYTVLTSIITLDRVSLKQKVV 239
Query 143 FSPTI 147
+P I
Sbjct 240 DAPEI 244
> Hs7661914
Length=389
Score = 107 bits (268), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 50/126 (39%), Positives = 82/126 (65%), Gaps = 0/126 (0%)
Query 24 AYRVAYAKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLKV 83
A+R Y KTV +G RLD++ LLR G+ Y D DL+ + AK +E+GGDW+RRN+LKV
Sbjct 123 AFRKTYDKTVALGHRLDIVFYLLRIGLFYMDNDLITRNTEKAKSLIEEGGDWDRRNRLKV 182
Query 84 IEALDFILSRNFTEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVVF 143
+ L + R+F +A++L L+ ++TF S ++ ++TFV Y ++++ +R L+E+V+
Sbjct 183 YQGLYCVAIRDFKQAAELFLDTVSTFTSYELMDYKTFVTYTVYVSMIALERPDLREKVIK 242
Query 144 SPTIAE 149
I E
Sbjct 243 GAEILE 248
> 7300859
Length=389
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 47/126 (37%), Positives = 78/126 (61%), Gaps = 0/126 (0%)
Query 24 AYRVAYAKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLKV 83
A+R Y KTV +G RLD++ L+R G+ Y D DL+ + + AK +E+GGDW+RRN+LKV
Sbjct 123 AFRKTYEKTVSLGHRLDIVFHLIRLGLFYLDHDLITRNIDKAKYLIEEGGDWDRRNRLKV 182
Query 84 IEALDFILSRNFTEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVVF 143
+ + + R+F A+ L+ ++TF S ++ + TFV Y +A++ R L+++V+
Sbjct 183 YQGVYSVAVRDFKAAATFFLDTVSTFTSYELMDYPTFVRYTVYVAMIALPRNELRDKVIK 242
Query 144 SPTIAE 149
I E
Sbjct 243 GSEIQE 248
> YPR108w
Length=429
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/118 (39%), Positives = 74/118 (62%), Gaps = 0/118 (0%)
Query 30 AKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLKVIEALDF 89
+K + G+++D++LT+ R G Y D+ VK++L +EKGGDWERRN+ K +
Sbjct 159 SKAISTGAKIDVMLTIARLGFFYNDQLYVKEKLEAVNSMIEKGGDWERRNRYKTYYGIHC 218
Query 90 ILSRNFTEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVVFSPTI 147
+ RNF EA+KLL++ LATF S + S+E+ YA++ L +R LK +V+ SP +
Sbjct 219 LAVRNFKEAAKLLVDSLATFTSIELTSYESIATYASVTGLFTLERTDLKSKVIDSPEL 276
> CE03368
Length=410
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 76/127 (59%), Gaps = 0/127 (0%)
Query 23 EAYRVAYAKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLK 82
+AY Y KTVG+G R+D++ ++R G+ + D L+ K + AK+ ME+GGDWER+N+L+
Sbjct 136 KAYTATYEKTVGMGYRIDVVFAMIRVGLFFLDHHLINKFITKAKELMEQGGDWERKNRLR 195
Query 83 VIEALDFILSRNFTEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVV 142
EAL + R+F A+ L LE + TF S ++++E + Y + DR L+ +V+
Sbjct 196 SYEALYRMSVRDFAGAADLFLEAVPTFGSYELMTYENLILYTVITTTFALDRPDLRTKVI 255
Query 143 FSPTIAE 149
+ E
Sbjct 256 RCNEVQE 262
> SPBC582.07c
Length=409
Score = 78.2 bits (191), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 67/112 (59%), Gaps = 0/112 (0%)
Query 36 GSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLKVIEALDFILSRNF 95
G ++D++ + +R VY D +V + L K +EKGGDWER+N+LK + + + RNF
Sbjct 146 GMKIDVLFSKIRLAYVYADMRVVGQLLEKLKPLIEKGGDWERKNRLKAYQGIYLMSIRNF 205
Query 96 TEASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVVFSPTI 147
+ A+ LLL+ ++TF S ++ + V YA + + DR +K ++V SP +
Sbjct 206 SGAADLLLDCMSTFSSTELLPYYDVVRYAVISGAISLDRVDVKTKIVDSPEV 257
> ECU10g0190
Length=381
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 62/105 (59%), Gaps = 0/105 (0%)
Query 38 RLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKLKVIEALDFILSRNFTE 97
++D+ L +R G++ +R +V++ + +A E+G DW+RRN+ KV + + ++ R F E
Sbjct 137 KMDIFLCKIRMGLIVGNRKVVEESIEVADDIYERGCDWDRRNRYKVYKGMFKMMRRKFKE 196
Query 98 ASKLLLEVLATFPSCGIISFETFVFYAALLALLCCDRQTLKEQVV 142
A L E+L +F S +IS+ V Y LL +R+ ++ +++
Sbjct 197 AGILFSEILPSFESSEVISYSRAVRYMIFCGLLRFERREIETRIL 241
> Hs8567388
Length=1201
Score = 34.7 bits (78), Expect = 0.063, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 53 KDRDLVKKQLFLAKQEMEKGGDWERRNKLKVIEALDFIL 91
+DR+ V ++L + QEM+K ERRNK ++AL++ L
Sbjct 46 QDRNRVSEELIMVVQEMKKYFPSERRNKPSTLDALNYAL 84
> At4g33300
Length=855
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Query 40 DLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRN 79
DL L L G V + +K+L + K+E++ GDWER N
Sbjct 509 DLALHLSNAGKVNR-----RKRLLMPKRELDLPGDWERNN 543
> CE03494
Length=832
Score = 28.5 bits (62), Expect = 4.3, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 10/63 (15%)
Query 67 QEMEKGGDWERRNKLKVIEALDFILSRNFT---EASKLLLEVLATFPSCGI-------IS 116
+E + G +R N+L I+ ++F++S +FT + + L+T PS I I
Sbjct 521 EEYKHGEGLQRVNELSAIDPMEFVISADFTPFEDRQMTYMHRLSTAPSSTIRKKHGKAIY 580
Query 117 FET 119
FET
Sbjct 581 FET 583
> CE27332
Length=530
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 31/69 (44%), Gaps = 0/69 (0%)
Query 22 REAYRVAYAKTVGIGSRLDLILTLLRTGIVYKDRDLVKKQLFLAKQEMEKGGDWERRNKL 81
+ A V + T G G + I L G RDL +K + L + + G+W+ R +
Sbjct 251 QRAQAVGFTDTSGDGQTVYRIKNHLPIGSYLMKRDLYEKHIRLREFCCTERGNWDLRESI 310
Query 82 KVIEALDFI 90
++ A+ I
Sbjct 311 SLVPAMSRI 319
> Hs19913396
Length=915
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 5/62 (8%)
Query 14 QNWRHTPRREA-YRVAYAKTVG-IGSRLDL--ILTLLRTGIVYKDRDLVKKQLFLAK-QE 68
+NW H+ +REA R A A+ G + S D+ + LLR+ Y++R L++ + + QE
Sbjct 63 ENWLHSQQREAEQRAALARLAGQLESMNDVEELTALLRSAGEYEERKLIRAAIRRVRAQE 122
Query 69 ME 70
+E
Sbjct 123 IE 124
> Hs19913394
Length=917
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 5/62 (8%)
Query 14 QNWRHTPRREA-YRVAYAKTVG-IGSRLDL--ILTLLRTGIVYKDRDLVKKQLFLAK-QE 68
+NW H+ +REA R A A+ G + S D+ + LLR+ Y++R L++ + + QE
Sbjct 63 ENWLHSQQREAEQRAALARLAGQLESMNDVEELTALLRSAGEYEERKLIRAAIRRVRAQE 122
Query 69 ME 70
+E
Sbjct 123 IE 124
> Hs19913398
Length=940
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 5/62 (8%)
Query 14 QNWRHTPRREA-YRVAYAKTVG-IGSRLDL--ILTLLRTGIVYKDRDLVKKQLFLAK-QE 68
+NW H+ +REA R A A+ G + S D+ + LLR+ Y++R L++ + + QE
Sbjct 63 ENWLHSQQREAEQRAALARLAGQLESMNDVEELTALLRSAGEYEERKLIRAAIRRVRAQE 122
Query 69 ME 70
+E
Sbjct 123 IE 124
> At1g64270
Length=441
Score = 27.3 bits (59), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Query 46 LRTGIVYKDRDLVKKQ-LFLAKQEMEKGGDWERRNKLKVIE 85
LR G+ +KDRD +KK + +EK GD + +KL+ E
Sbjct 194 LRVGLCFKDRDELKKAGTTHSNSRVEKFGDMLKTSKLRAAE 234
Lambda K H
0.324 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40