bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5426_orf3
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
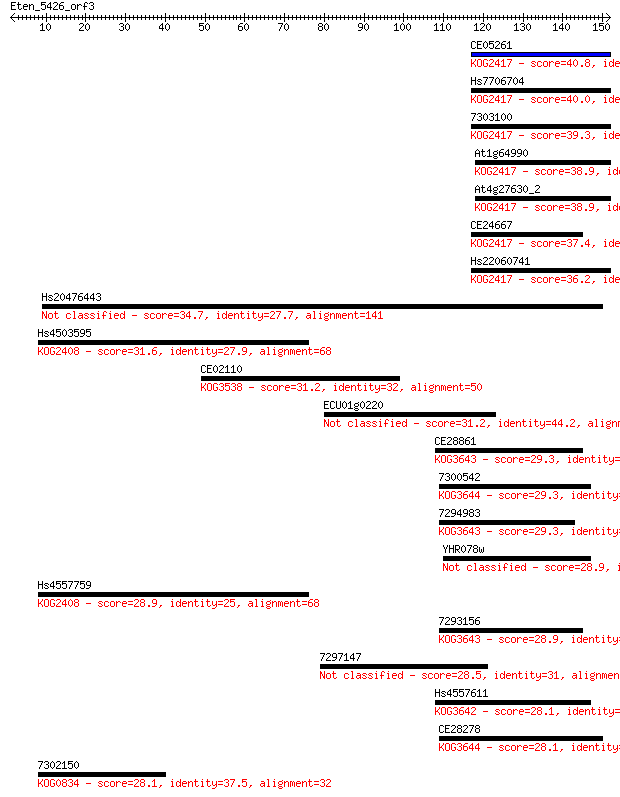
CE05261 40.8 0.001
Hs7706704 40.0 0.002
7303100 39.3 0.003
At1g64990 38.9 0.004
At4g27630_2 38.9 0.004
CE24667 37.4 0.011
Hs22060741 36.2 0.025
Hs20476443 34.7 0.062
Hs4503595 31.6 0.58
CE02110 31.2 0.75
ECU01g0220 31.2 0.87
CE28861 29.3 2.7
7300542 29.3 2.9
7294983 29.3 3.4
YHR078w 28.9 3.6
Hs4557759 28.9 3.9
7293156 28.9 4.2
7297147 28.5 5.4
Hs4557611 28.1 5.8
CE28278 28.1 5.9
7302150 28.1 6.2
> CE05261
Length=434
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 117 ERLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
E++++ VGV GVTV++ L+GFG+VN PY + F+
Sbjct 145 EQVISRVGVIGVTVMAVLSGFGAVNAPYSYMTIFM 179
> Hs7706704
Length=455
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 117 ERLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
E+L++ VGV GVT+++ L+GFG+VN PY ++ FL
Sbjct 144 EQLISRVGVIGVTLMALLSGFGAVNCPYTYMSYFL 178
> 7303100
Length=441
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 117 ERLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
E+ ++ + V GVTV++ L+GFG+VNYPY +++ F+
Sbjct 131 EQGVSRISVIGVTVMAILSGFGAVNYPYTSMSYFI 165
> At1g64990
Length=465
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 118 RLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
+L++ +GV GVT+++ L+GFG+VN PY ++ F+
Sbjct 148 QLVSRIGVIGVTLMAVLSGFGAVNLPYSYISLFI 181
> At4g27630_2
Length=418
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 118 RLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
+L++ +GV GVT+++ L+GFG+VN PY ++ F+
Sbjct 147 QLVSRIGVIGVTLMAVLSGFGAVNLPYSYISLFI 180
> CE24667
Length=465
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 24/28 (85%), Gaps = 0/28 (0%)
Query 117 ERLLAYVGVCGVTVVSALAGFGSVNYPY 144
E++++ VGV GVT+++ L+GFG+VN PY
Sbjct 148 EQVISRVGVVGVTIMAILSGFGAVNAPY 175
> Hs22060741
Length=559
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 117 ERLLAYVGVCGVTVVSALAGFGSVNYPYRNVATFL 151
E+L+++VGV GVT+++ L+GFG+V Y ++ FL
Sbjct 119 EQLISWVGVIGVTLMALLSGFGAVYCTYTLMSYFL 153
> Hs20476443
Length=633
Score = 34.7 bits (78), Expect = 0.062, Method: Composition-based stats.
Identities = 39/144 (27%), Positives = 58/144 (40%), Gaps = 20/144 (13%)
Query 9 LPVSIIHNALAPSADSFRIADWAAKFPQMQRLQTRAAALSRLAAVS-TLASSSGNGVLSK 67
+P S +HN + SF ++ F + L A +A +LAS N VL
Sbjct 1 MPNSSVHN----NGKSFINSETIEDFQKKNNLYPHRTAFPSVALKGHSLASVFKNSVLRS 56
Query 68 LGKLSPRWREWVAVAVCSAVVLPFLWFCFCQSGRLLHLDSEALAELSYTERLLAYVGVCG 127
LG S + AV CF +SGR LH S+ L + + ++ G G
Sbjct 57 LGAASNPFPYKAAV-------------CFAESGRNLHSSSQQLLPFAASPATCSFSGEKG 103
Query 128 VTVVSA--LAGFGSVNYPYRNVAT 149
+ VS L VN P + V++
Sbjct 104 LLPVSENDLESTSKVNIPVKVVSS 127
> Hs4503595
Length=715
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 8 VLPVSIIHNALAPSADSFRIADWAAKFPQMQRLQTRAAALSRLAAVSTLASSSGNGVLSK 67
+LP +H+ D + + +A+ P TR+ +LAA+ TL N + ++
Sbjct 348 LLPFDNLHD------DPCLLTNRSARIPCFLAGDTRSTETPKLAAMHTLFMREHNRLATE 401
Query 68 LGKLSPRW 75
L +L+PRW
Sbjct 402 LRRLNPRW 409
> CE02110
Length=1444
Score = 31.2 bits (69), Expect = 0.75, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 49 RLAAVSTLASSSGNGVLSKLGKLSPRWREWVAVAVCSAVVLPFLWFCFCQ 98
R + + +S S +S+ G +PRW EW + + CS L FCQ
Sbjct 1108 RCKDLPSCSSISSGRTISENGFDAPRWSEWSSWSACSCFSLTSTRRRFCQ 1157
> ECU01g0220
Length=247
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 80 AVAVCSAVVLPFLW-FCFCQSGRLLHLDSEALAELSYTERLLAY 122
++ V S VV LW F + GR+L + EALA+LS+T + AY
Sbjct 22 SIYVVSMVVFTVLWSFGIEKKGRVLVIAVEALAQLSFTMSVAAY 65
> CE28861
Length=500
Score = 29.3 bits (64), Expect = 2.7, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 108 EALAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPY 144
++L +SY + + Y+ +C V V +AL + +VNY Y
Sbjct 275 QSLPRISYVKSIDIYLVMCFVFVFAALLEYAAVNYSY 311
> 7300542
Length=454
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 109 ALAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPYRN 146
+L +SYT+ + + GVC V AL F VNY R+
Sbjct 295 SLPPVSYTKAIDVWTGVCLTFVFGALLEFALVNYASRS 332
> 7294983
Length=606
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 109 ALAELSYTERLLAYVGVCGVTVVSALAGFGSVNY 142
AL ++SY + + Y+G C V V ++L + +V Y
Sbjct 320 ALPKISYVKSIDVYLGTCFVMVFASLLEYATVGY 353
> YHR078w
Length=552
Score = 28.9 bits (63), Expect = 3.6, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 110 LAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPYRN 146
+ YT+ +L + + GVTV+++L+G +V+ Y N
Sbjct 180 IGPFQYTKNILTRLSIGGVTVMASLSGLATVSSLYYN 216
> Hs4557759
Length=745
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 33/68 (48%), Gaps = 6/68 (8%)
Query 8 VLPVSIIHNALAPSADSFRIADWAAKFPQMQRLQTRAAALSRLAAVSTLASSSGNGVLSK 67
+LP +H+ D + + +A+ P TR++ + L ++ TL N + ++
Sbjct 376 LLPFDNLHD------DPCLLTNRSARIPCFLAGDTRSSEMPELTSMHTLLLREHNRLATE 429
Query 68 LGKLSPRW 75
L L+PRW
Sbjct 430 LKSLNPRW 437
> 7293156
Length=496
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 109 ALAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPY 144
+L +SY + + Y+ +C V V +AL + +VNY Y
Sbjct 309 SLPRISYVKAIDIYLVMCFVFVFAALLEYAAVNYTY 344
> 7297147
Length=1377
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 79 VAVAVCSAVVLPFLWFCFCQSGRLLHLDSEALAELSYTERLL 120
V +A S++ F ++CFC S + + L++L++ ER L
Sbjct 747 VVIAAISSLFFMFAYYCFCNSSQ----EGALLSKLNHLERSL 784
> Hs4557611
Length=467
Score = 28.1 bits (61), Expect = 5.8, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 108 EALAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPYRN 146
++L ++SY + +V VC + V SAL +G+++Y N
Sbjct 324 KSLPKVSYVTAMDLFVSVCFIFVFSALVEYGTLHYFVSN 362
> CE28278
Length=636
Score = 28.1 bits (61), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 109 ALAELSYTERLLAYVGVCGVTVVSALAGFGSVNYPYRNVAT 149
L +SYT+ + ++GVC + AL F VNY R T
Sbjct 306 KLPPVSYTKAIDVWIGVCLAFIFGALLEFALVNYAARKDMT 346
> 7302150
Length=381
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 18/32 (56%), Gaps = 2/32 (6%)
Query 8 VLPVSIIHNALAPSADSFRIADWAAKFPQMQR 39
++ V++IH LA F + DW + PQ QR
Sbjct 185 IIAVALIH--LASKLSKFTVQDWEGRQPQQQR 214
Lambda K H
0.326 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40