bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5409_orf1
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
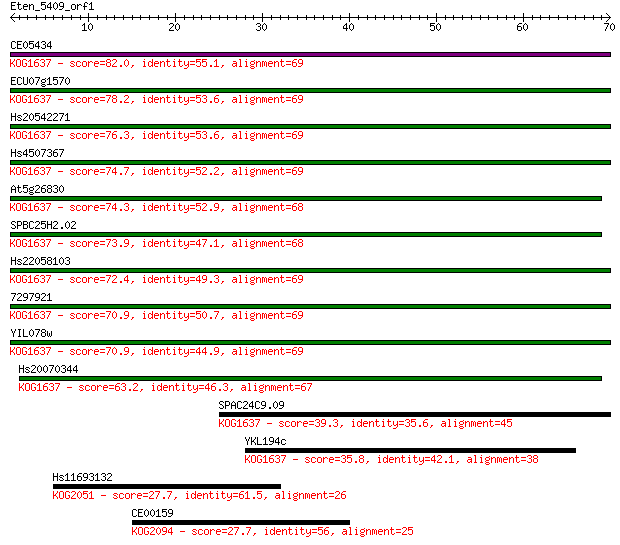
CE05434 82.0 2e-16
ECU07g1570 78.2 4e-15
Hs20542271 76.3 1e-14
Hs4507367 74.7 4e-14
At5g26830 74.3 6e-14
SPBC25H2.02 73.9 6e-14
Hs22058103 72.4 2e-13
7297921 70.9 6e-13
YIL078w 70.9 6e-13
Hs20070344 63.2 1e-10
SPAC24C9.09 39.3 0.002
YKL194c 35.8 0.023
Hs11693132 27.7 5.1
CE00159 27.7 5.8
> CE05434
Length=725
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ KQLKE+ L E+A +RDHR LG LFFF +S G AFW P+GA +YN L
Sbjct 297 YGISFPDSKQLKEWQKLQEEAAKRDHRKLGKEHDLFFFH-QLSPGSAFWYPKGAHIYNKL 355
Query 61 IEFIREEYQ 69
++FIR++Y+
Sbjct 356 VDFIRKQYR 364
> ECU07g1570
Length=640
Score = 78.2 bits (191), Expect = 4e-15, Method: Composition-based stats.
Identities = 37/69 (53%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
Y ++FP K + EYL E+AKERDHR +G L LFFF S G F+LP G +YN L
Sbjct 217 YAITFPSKGMMDEYLKRKEEAKERDHRKIGTELDLFFFS-KYSPGSCFFLPNGTTMYNTL 275
Query 61 IEFIREEYQ 69
IEF+REEY+
Sbjct 276 IEFLREEYR 284
> Hs20542271
Length=723
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 37/69 (53%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K LKE+ E+AK RDHR +G + L+FF +S G F+LP+GA +YN L
Sbjct 298 YGISFPDPKMLKEWEKFQEEAKNRDHRKIGRDQELYFFH-ELSPGSCFFLPKGAYIYNAL 356
Query 61 IEFIREEYQ 69
IEFIR EY+
Sbjct 357 IEFIRSEYR 365
> Hs4507367
Length=712
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 48/69 (69%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K LKE+ E+AK RDHR +G + L+FF +S G F+LP+G +YN L
Sbjct 286 YGISFPDPKMLKEWEKFQEEAKNRDHRKIGRDQELYFFH-ELSPGSCFFLPKGVYIYNAL 344
Query 61 IEFIREEYQ 69
IEFIR EY+
Sbjct 345 IEFIRSEYR 353
> At5g26830
Length=676
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 50/68 (73%), Gaps = 1/68 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+S+P++KQLK+YL LE+AK+ DHRLLG L FF +S G F+LP G +VYN L
Sbjct 259 YGISYPDQKQLKKYLQFLEEAKKYDHRLLGQKQEL-FFSHQLSPGSYFFLPLGTRVYNRL 317
Query 61 IEFIREEY 68
++FI+ +Y
Sbjct 318 MDFIKNQY 325
> SPBC25H2.02
Length=703
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 49/68 (72%), Gaps = 1/68 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ KQ++EY L +A +RDHR +G + LFFF +S G F+LP GA++YN L
Sbjct 266 YGISFPDNKQMQEYKTFLAEAAKRDHRKIGRDQELFFFN-EISPGSCFFLPHGARIYNTL 324
Query 61 IEFIREEY 68
++++R +Y
Sbjct 325 LKYMRYQY 332
> Hs22058103
Length=707
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K ++++ E+AK RDHR +G LFFF +S G F+LP GA +YN L
Sbjct 282 YGISFPDNKMMRDWEKFQEEAKNRDHRKIGKEQELFFFH-DLSPGSCFFLPRGAFIYNTL 340
Query 61 IEFIREEYQ 69
+FIREEY
Sbjct 341 TDFIREEYH 349
> 7297921
Length=690
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 47/69 (68%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ KQLKE+ L E+A +RDHR +G LFFF +S G F+ P GA +YN L
Sbjct 267 YGISFPDPKQLKEWEKLQEEAAKRDHRKIGREQELFFFH-ELSPGSCFFQPRGAHIYNTL 325
Query 61 IEFIREEYQ 69
+ FI+ EY+
Sbjct 326 MGFIKAEYR 334
> YIL078w
Length=734
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 44/69 (63%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+KK + +L L +A RDHR +G LF F +S G FWLP G ++YN L
Sbjct 305 YGISFPDKKLMDAHLKFLAEASMRDHRKIGKEQELFLFN-EMSPGSCFWLPHGTRIYNTL 363
Query 61 IEFIREEYQ 69
++ +R EY+
Sbjct 364 VDLLRTEYR 372
> Hs20070344
Length=718
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 2 GVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLI 61
G+SFP + L+ + E+A+ RDHR +G LFFF +S G F+LP G +VYN L+
Sbjct 278 GISFPTTELLRVWEAWREEAELRDHRRIGKEQELFFFH-ELSPGSCFFLPRGTRVYNALV 336
Query 62 EFIREEY 68
FIR EY
Sbjct 337 AFIRAEY 343
> SPAC24C9.09
Length=473
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Query 25 DHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLIEFIREEYQ 69
DHR + L+ ++ G F+LP G ++YN L++F+R +YQ
Sbjct 47 DHRTIAARQKLYT-TSILTPGSIFFLPHGTRIYNRLVDFLRAQYQ 90
> YKL194c
Length=462
Score = 35.8 bits (81), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query 28 LLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLIEFIR 65
++ LF +P +S G F+LP GAK++N LIEF++
Sbjct 41 MVSQRQDLFMTDP-LSPGSMFFLPNGAKIFNKLIEFMK 77
> Hs11693132
Length=1272
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 16/31 (51%), Positives = 18/31 (58%), Gaps = 5/31 (16%)
Query 6 PEKKQ-----LKEYLDLLEKAKERDHRLLGN 31
PEK+Q LKEY L K +RDHR L N
Sbjct 346 PEKQQPFQNLLKEYFTSLTKHLKRDHRELQN 376
> CE00159
Length=518
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 15 LDLLEKAKERDHRLLGNNLSLFFFE 39
L+LLEK K ++ RLLG LS FE
Sbjct 452 LELLEKEKGKEIRLLGVRLSQLIFE 476
Lambda K H
0.319 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200381448
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40