bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5341_orf1
Length=116
Score E
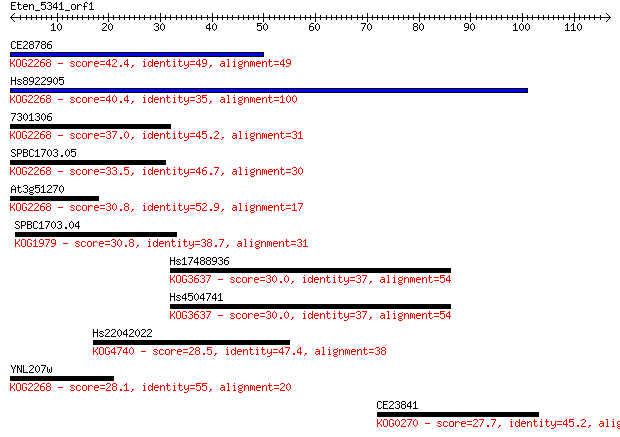
Sequences producing significant alignments: (Bits) Value
CE28786 42.4 2e-04
Hs8922905 40.4 7e-04
7301306 37.0 0.009
SPBC1703.05 33.5 0.11
At3g51270 30.8 0.55
SPBC1703.04 30.8 0.72
Hs17488936 30.0 1.1
Hs4504741 30.0 1.2
Hs22042022 28.5 3.3
YNL207w 28.1 4.5
CE23841 27.7 4.7
> CE28786
Length=562
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 30/60 (50%), Gaps = 11/60 (18%)
Query 1 YFDRDVQCIKRLFERKFLVEVTTVPKFDEV-----------VEGRETKLSSAADGALDEG 49
YFDRDV C++ F+RKF E PKFDEV G K++ + A DEG
Sbjct 294 YFDRDVTCVRTFFKRKFDYESEDWPKFDEVERKGNMDVLLEASGFTKKMALDLNKAYDEG 353
> Hs8922905
Length=552
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 49/109 (44%), Gaps = 15/109 (13%)
Query 1 YFDRDVQCIKRLFERKFLVEVTTVPKFDEVVEGRETKLSSAADGALDEGRAE-------G 53
YFDRDV+CIK F ++F E P F ++ + +A G E +A+ G
Sbjct 261 YFDRDVKCIKDFFMKRFSYESELFPTFKDIRREDTLDVEVSASGYTKEMQADDELLHPLG 320
Query 54 AADG--EESSCNLISVSDDLDEHQAELLEGALAARQNNETESSCSDESE 100
D E + S SD E+ E A R NE+E +C +ESE
Sbjct 321 PDDKNIETKEGSEFSFSD------GEVAEKAEVYRSENESERNCLEESE 363
> 7301306
Length=538
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 1 YFDRDVQCIKRLFERKFLVEVTTVPKFDEVV 31
+F+RDV C++ +F RKF E PKF ++V
Sbjct 261 FFERDVNCVREMFRRKFGYESEDYPKFSDLV 291
> SPBC1703.05
Length=336
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 21/30 (70%), Gaps = 1/30 (3%)
Query 1 YFDRDVQCIKRLFERKFLVEVTTVPKFDEV 30
YFDRDVQCI + FE+ + + VP F+++
Sbjct 253 YFDRDVQCIVQYFEKNYQYK-GDVPNFEDI 281
> At3g51270
Length=472
Score = 30.8 bits (68), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 9/17 (52%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 1 YFDRDVQCIKRLFERKF 17
YFDRD++CI + F ++F
Sbjct 270 YFDRDIECIFKFFRKRF 286
> SPBC1703.04
Length=684
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 2 FDRDVQCIKRLFERKFLVEVTTVPKFDEVVE 32
F R V C K++FE K + ++T++P+ V E
Sbjct 652 FRRRVICPKKVFEEKCIYQITSLPRLYNVFE 682
> Hs17488936
Length=1167
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 32 EGRETKLSSAADGALDEGRAEGAADGEESSCNLISVSDDLDEHQAELLEGALAA 85
EGRETK + A A EG ++ G + L+ V D + H E L AL A
Sbjct 239 EGRETKTAQAIMVACTEGFSQSHG-GRPEAARLLVVVTDGESHDGEELPAALKA 291
> Hs4504741
Length=1167
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 32 EGRETKLSSAADGALDEGRAEGAADGEESSCNLISVSDDLDEHQAELLEGALAA 85
EGRETK + A A EG ++ G + L+ V D + H E L AL A
Sbjct 239 EGRETKTAQAIMVACTEGFSQSHG-GRPEAARLLVVVTDGESHDGEELPAALKA 291
> Hs22042022
Length=330
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 19/38 (50%), Gaps = 4/38 (10%)
Query 17 FLVEVTTVPKFDEVVEGRETKLSSAADGALDEGRAEGA 54
FL VPK GRE +LSS A GA E A GA
Sbjct 236 FLCNTLVVPK----ASGREQELSSPASGASPEAPAAGA 269
> YNL207w
Length=425
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 1 YFDRDVQCIKRLFERKFLVE 20
YF RDV CI+R F++K E
Sbjct 268 YFQRDVDCIRRFFKKKLKYE 287
> CE23841
Length=476
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 72 DEHQAELLEGALAARQNNETESSCSDESETE 102
DE EL+EGA+ +NETE +ES+ E
Sbjct 38 DEKLKELIEGAIPESTDNETELDDDEESDNE 68
Lambda K H
0.305 0.124 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40