bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5328_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
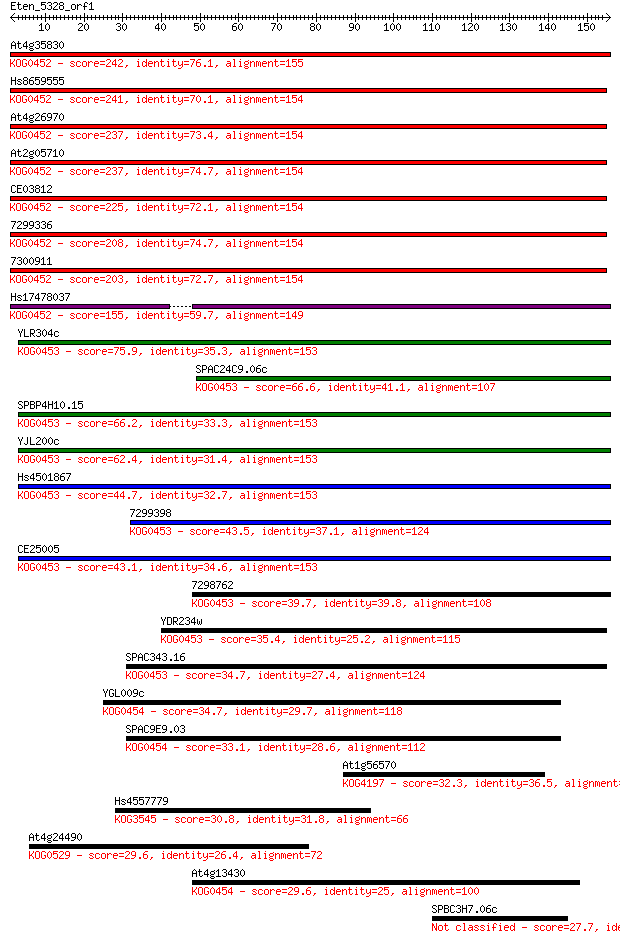
At4g35830 242 2e-64
Hs8659555 241 4e-64
At4g26970 237 6e-63
At2g05710 237 8e-63
CE03812 225 2e-59
7299336 208 3e-54
7300911 203 1e-52
Hs17478037 155 4e-38
YLR304c 75.9 3e-14
SPAC24C9.06c 66.6 2e-11
SPBP4H10.15 66.2 2e-11
YJL200c 62.4 3e-10
Hs4501867 44.7 8e-05
7299398 43.5 2e-04
CE25005 43.1 2e-04
7298762 39.7 0.002
YDR234w 35.4 0.049
SPAC343.16 34.7 0.073
YGL009c 34.7 0.080
SPAC9E9.03 33.1 0.20
At1g56570 32.3 0.34
Hs4557779 30.8 1.2
At4g24490 29.6 2.2
At4g13430 29.6 2.2
SPBC3H7.06c 27.7 9.0
> At4g35830
Length=898
Score = 242 bits (617), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 118/155 (76%), Positives = 131/155 (84%), Gaps = 0/155 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVDLA MRDAM LGGD INPLV VDLV+DHSVQVD +RSE+A+Q N+E E +RNKER
Sbjct 98 VVDLACMRDAMNNLGGDSNKINPLVPVDLVIDHSVQVDVARSENAVQANMELEFQRNKER 157
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNG 120
FAFLKWGS AF+NM +VPPGSGIVHQVNLE+LA V + GLLYPDSVVGTDSHTTM++G
Sbjct 158 FAFLKWGSNAFHNMLVVPPGSGIVHQVNLEYLARVVFNTNGLLYPDSVVGTDSHTTMIDG 217
Query 121 LGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
LGV GWGVGGIEAEA MLGQ +SMVLP VVG LT
Sbjct 218 LGVAGWGVGGIEAEATMLGQPMSMVLPGVVGFKLT 252
> Hs8659555
Length=889
Score = 241 bits (615), Expect = 4e-64, Method: Compositional matrix adjust.
Identities = 108/154 (70%), Positives = 131/154 (85%), Gaps = 0/154 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVD AAMRDA+ +LGGDP INP+ DLV+DHS+QVD++R +LQ+N + E ERN+ER
Sbjct 94 VVDFAAMRDAVKKLGGDPEKINPVCPADLVIDHSIQVDFNRRADSLQKNQDLEFERNRER 153
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNG 120
F FLKWGS+AF+NMRI+PPGSGI+HQVNLE+LA V D+ G YPDS+VGTDSHTTM++G
Sbjct 154 FEFLKWGSQAFHNMRIIPPGSGIIHQVNLEYLARVVFDQDGYYYPDSLVGTDSHTTMIDG 213
Query 121 LGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
LG+LGWGVGGIEAEAVMLGQ ISMVLP+V+G L
Sbjct 214 LGILGWGVGGIEAEAVMLGQPISMVLPQVIGYRL 247
> At4g26970
Length=907
Score = 237 bits (605), Expect = 6e-63, Method: Compositional matrix adjust.
Identities = 113/154 (73%), Positives = 127/154 (82%), Gaps = 0/154 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
+VDLA+MRDA+ LG DP INPLV VDLVVDHS+QVD++RSE A Q+NLE E +RNKER
Sbjct 107 LVDLASMRDAVKNLGSDPSKINPLVPVDLVVDHSIQVDFARSEDAAQKNLELEFKRNKER 166
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNG 120
F FLKWGS AF NM +VPPGSGIVHQVNLE+L V + G LYPDSVVGTDSHTTM++G
Sbjct 167 FTFLKWGSTAFQNMLVVPPGSGIVHQVNLEYLGRVVFNSKGFLYPDSVVGTDSHTTMIDG 226
Query 121 LGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
LGV GWGVGGIEAEA MLGQ +SMVLP VVG L
Sbjct 227 LGVAGWGVGGIEAEAAMLGQPMSMVLPGVVGFKL 260
> At2g05710
Length=898
Score = 237 bits (604), Expect = 8e-63, Method: Compositional matrix adjust.
Identities = 115/154 (74%), Positives = 128/154 (83%), Gaps = 0/154 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVDLA MRDAM +LG D INPLV VDLV+DHSVQVD +RSE+A+Q N+E E +RNKER
Sbjct 98 VVDLACMRDAMNKLGSDSNKINPLVPVDLVIDHSVQVDVARSENAVQANMELEFQRNKER 157
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNG 120
FAFLKWGS AF NM +VPPGSGIVHQVNLE+L V + GLLYPDSVVGTDSHTTM++G
Sbjct 158 FAFLKWGSTAFQNMLVVPPGSGIVHQVNLEYLGRVVFNTKGLLYPDSVVGTDSHTTMIDG 217
Query 121 LGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
LGV GWGVGGIEAEA MLGQ +SMVLP VVG L
Sbjct 218 LGVAGWGVGGIEAEATMLGQPMSMVLPGVVGFKL 251
> CE03812
Length=887
Score = 225 bits (574), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 111/155 (71%), Positives = 129/155 (83%), Gaps = 1/155 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVDLAAMRDA+ +G DP INP+ VDLV+DHSVQVD+ + AL +N E ERN+ER
Sbjct 92 VVDLAAMRDAVQNMGADPAKINPVCPVDLVIDHSVQVDHYGNLEALAKNQSIEFERNRER 151
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLA-SVTMDKGGLLYPDSVVGTDSHTTMVN 119
F FLKWGSKAF+N+ IVPPGSGIVHQVNLE+LA +V + K G+LYPDSVVGTDSHTTM++
Sbjct 152 FNFLKWGSKAFDNLLIVPPGSGIVHQVNLEYLARTVFVGKDGVLYPDSVVGTDSHTTMID 211
Query 120 GLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
G GVLGWGVGGIEAEAVMLGQ ISMV+PEV+G L
Sbjct 212 GSGVLGWGVGGIEAEAVMLGQPISMVIPEVIGYEL 246
> 7299336
Length=899
Score = 208 bits (529), Expect = 3e-54, Method: Composition-based stats.
Identities = 115/159 (72%), Positives = 128/159 (80%), Gaps = 5/159 (3%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVD AAMRDA+ LGGDP INP+ DLV+DHSVQVD++R+ AL +N E ERNKER
Sbjct 98 VVDFAAMRDAVLDLGGDPEKINPICPADLVIDHSVQVDFARAPDALAKNQSLEFERNKER 157
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTM-----DKGGLLYPDSVVGTDSHT 115
F FLKWG+KAFNNM IVPPGSGIVHQVNLE+LA V D +LYPDSVVGTDSHT
Sbjct 158 FTFLKWGAKAFNNMLIVPPGSGIVHQVNLEYLARVVFENDATDGSKILYPDSVVGTDSHT 217
Query 116 TMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
TM+NGLGVLGWGVGGIEAEAVMLGQ ISM+LPEV+G L
Sbjct 218 TMINGLGVLGWGVGGIEAEAVMLGQSISMLLPEVIGYKL 256
> 7300911
Length=902
Score = 203 bits (516), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 112/160 (70%), Positives = 128/160 (80%), Gaps = 6/160 (3%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKER 60
VVD AAMRDA+ LGG+P INP+ DLV+DHSVQVD+ RS AL +N E +RNKER
Sbjct 100 VVDFAAMRDAVRELGGNPEKINPICPADLVIDHSVQVDFVRSSDALTKNESLEFQRNKER 159
Query 61 FAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASV------TMDKGGLLYPDSVVGTDSH 114
F FLKWG++AF+NM IVPPGSGIVHQVNLE+LA V + D +LYPDSVVGTDSH
Sbjct 160 FTFLKWGARAFDNMLIVPPGSGIVHQVNLEYLARVVFESDSSADGSKILYPDSVVGTDSH 219
Query 115 TTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
TTM+NGLGVLGWGVGGIEAEAVMLGQ ISM+LPEV+G L
Sbjct 220 TTMINGLGVLGWGVGGIEAEAVMLGQSISMLLPEVIGYRL 259
> Hs17478037
Length=963
Score = 155 bits (391), Expect = 4e-38, Method: Composition-based stats.
Identities = 67/108 (62%), Positives = 84/108 (77%), Gaps = 0/108 (0%)
Query 48 RNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDS 107
+N E E RN+ER F KW S+ F N+ ++PPG+G+ HQ+NLE+L+ V ++ LL+PDS
Sbjct 216 KNQEVEFGRNRERLQFFKWSSRVFKNVAVIPPGTGMAHQINLEYLSRVVFEEKDLLFPDS 275
Query 108 VVGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
VVGTDSH TMVNGLG+LGWGVGGIE EAVMLG +S+ LPEVVG LT
Sbjct 276 VVGTDSHITMVNGLGILGWGVGGIETEAVMLGLPVSLTLPEVVGCELT 323
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 22/41 (53%), Positives = 31/41 (75%), Gaps = 0/41 (0%)
Query 1 VVDLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSR 41
+VD AAMR+A+ LGGDP ++P DL VDHS+Q+D+S+
Sbjct 96 MVDFAAMREAVKTLGGDPEKVHPACPTDLTVDHSLQIDFSK 136
> YLR304c
Length=778
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 54/161 (33%), Positives = 85/161 (52%), Gaps = 27/161 (16%)
Query 3 DLAAMRDAMARLG-------GDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEME 55
D A +DA A++ G P++ P V + DH +Q ++ +++L++ ++
Sbjct 90 DRVACQDATAQMAILQFMSAGLPQVAKP---VTVHCDHLIQ-----AQVGGEKDLKRAID 141
Query 56 RNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDS-VVGTDSH 114
NKE + FL + +N M PGSGI+HQ+ LE A +P + ++GTDSH
Sbjct 142 LNKEVYDFLASATAKYN-MGFWKPGSGIIHQIVLENYA----------FPGALIIGTDSH 190
Query 115 TTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
T GLG L GVGG +A VM G+ + P+++GV LT
Sbjct 191 TPNAGGLGQLAIGVGGADAVDVMAGRPWELKAPKILGVKLT 231
> SPAC24C9.06c
Length=778
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 44/107 (41%), Positives = 59/107 (55%), Gaps = 10/107 (9%)
Query 49 NLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSV 108
+LE+ NKE + FL+ +N + PGSGI+HQ+ LE A GGLL
Sbjct 135 DLERANVTNKEVYDFLQTACAKYN-IGFWRPGSGIIHQIVLENYAF----PGGLL----- 184
Query 109 VGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
+GTDSHT GLG++ GVGG +A VM + P+V+GV LT
Sbjct 185 IGTDSHTPNAGGLGMVAIGVGGADAVDVMANLPWELKCPKVIGVKLT 231
> SPBP4H10.15
Length=905
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 51/162 (31%), Positives = 80/162 (49%), Gaps = 29/162 (17%)
Query 3 DLAAMRDAMARL--------GGDPRLINPLVDVD-LVVDHSVQVDYSRSESALQRNLEKE 53
D AM+DA A++ G + +I + D L+V H ++
Sbjct 94 DRVAMQDASAQMALLQFMTCGLEKTMIPASIHCDHLIVGHR----------GANSDIPDS 143
Query 54 MERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDS 113
+ NKE F FL+ +K + ++ PGSGI+HQ+ LE A+ GG++ +GTDS
Sbjct 144 IANNKEIFDFLQSAAKKYG-IQFWGPGSGIIHQIVLENYAA----PGGMM-----LGTDS 193
Query 114 HTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
HT GLG++ GVGG +A M + P+++GV+LT
Sbjct 194 HTPNAGGLGMIAIGVGGADAVDAMTNTPWELKAPKIIGVNLT 235
> YJL200c
Length=789
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 48/154 (31%), Positives = 78/154 (50%), Gaps = 13/154 (8%)
Query 3 DLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDY-SRSESALQRNLEKEMERNKERF 61
D AM+DA A++ + L V S+ D+ + ++L + N+E F
Sbjct 91 DRVAMQDASAQMALLQFMTTGLNQTS--VPASIHCDHLIVGKDGETKDLPSSIATNQEVF 148
Query 62 AFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNGL 121
FL+ +K + ++ PGSGI+HQ+ LE ++ GL+ ++GTDSHT GL
Sbjct 149 DFLESCAKRYG-IQFWGPGSGIIHQIVLENFSA-----PGLM----MLGTDSHTPNAGGL 198
Query 122 GVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
G + GVGG +A + G + P+++GV LT
Sbjct 199 GAIAIGVGGADAVDALTGTPWELKAPKILGVKLT 232
> Hs4501867
Length=780
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 50/155 (32%), Positives = 78/155 (50%), Gaps = 15/155 (9%)
Query 3 DLAAMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDY-SRSESALQRNLEKEMERNKERF 61
D AM+DA A++ + + L V V ++ D+ ++ +++L + + N+E +
Sbjct 94 DRVAMQDATAQMAMLQFISSGLSKV--AVPSTIHCDHLIEAQVGGEKDLRRAKDINQEVY 151
Query 62 AFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSV-VGTDSHTTMVNG 120
FL + + PGSGI+HQ+ LE A YP + +GTDSHT G
Sbjct 152 NFLATAGAKYG-VGFWKPGSGIIHQIILENYA----------YPGVLLIGTDSHTPNGGG 200
Query 121 LGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
LG + GVGG +A VM G + P+V+GV LT
Sbjct 201 LGGICIGVGGADAVDVMAGIPWELKCPKVIGVKLT 235
> 7299398
Length=769
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 46/124 (37%), Positives = 62/124 (50%), Gaps = 15/124 (12%)
Query 32 DHSVQVDYSRSESALQRNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEF 91
DH ++ S ++L + + NKE + FL + N+ PGSGI+HQ+ LE
Sbjct 111 DHLIEAQISGD-----KDLARAKDLNKEVYDFLSSACAKY-NLGFWKPGSGIIHQIILEN 164
Query 92 LASVTMDKGGLLYPDSVVGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVG 151
A GLL ++GTDSHT GLG L GVGG +A VM + P V+G
Sbjct 165 YAF-----PGLL----MIGTDSHTPNGGGLGCLCVGVGGADAVDVMANIPWELKCPTVIG 215
Query 152 VHLT 155
HLT
Sbjct 216 CHLT 219
> CE25005
Length=777
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 53/160 (33%), Positives = 77/160 (48%), Gaps = 25/160 (15%)
Query 3 DLAAMRDAMARLG-------GDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEME 55
D AM+DA A++ G P+ P + DH ++ ++ ++L + +
Sbjct 91 DRVAMQDATAQMAMLQFISSGLPKTAVPST---IHCDHLIE-----AQKGGAQDLARAKD 142
Query 56 RNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHT 115
NKE F FL + + PGSGI+HQ+ LE A GLL ++GTDSHT
Sbjct 143 LNKEVFNFLATAGSKYG-VGFWKPGSGIIHQIILENYAF-----PGLL----LIGTDSHT 192
Query 116 TMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
GLG L GVGG +A VM + P+V+G+ LT
Sbjct 193 PNGGGLGGLCIGVGGADAVDVMADIPWELKCPKVIGIKLT 232
> 7298762
Length=683
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 43/108 (39%), Positives = 59/108 (54%), Gaps = 10/108 (9%)
Query 48 RNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDS 107
++L + + NKE + FL + A + PGSGI+HQ+ LE A GLL
Sbjct 41 KDLARAKDLNKEVYDFLA-STCAKYGLGFWKPGSGIIHQIILENYAF-----PGLL---- 90
Query 108 VVGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHLT 155
++GTDSHT GLG L GVGG +A VM + P+V+GV+LT
Sbjct 91 MIGTDSHTPNGGGLGGLCIGVGGADAVDVMADIPWELKCPKVIGVNLT 138
> YDR234w
Length=693
Score = 35.4 bits (80), Expect = 0.049, Method: Composition-based stats.
Identities = 29/116 (25%), Positives = 54/116 (46%), Gaps = 11/116 (9%)
Query 40 SRSESALQRNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDK 99
S+ + L +++ + E+N ++ ++ +K +++ P G GI HQ+ +E
Sbjct 79 SQIVTTLDHDIQNKSEKNLTKYKNIENFAKK-HHIDHYPAGRGIGHQIMIE--------- 128
Query 100 GGLLYP-DSVVGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEVVGVHL 154
G +P + V +DSH+ GLG LG + +A A+ +P V V L
Sbjct 129 EGYAFPLNMTVASDSHSNTYGGLGSLGTPIVRTDAAAIWATGQTWWQIPPVAQVEL 184
> SPAC343.16
Length=721
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 54/125 (43%), Gaps = 26/125 (20%)
Query 31 VDHSVQVDYSRSESALQRNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLE 90
+DH VQ ++SE+ L++ +N E FA + P G GI HQ+ +E
Sbjct 125 LDHDVQ---NKSEANLRK------YKNIESFA-------KGQGIDFYPAGRGIGHQIMVE 168
Query 91 FLASVTMDKGGLLYPDSV-VGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLPEV 149
G P S+ V +DSH+ G+G LG + +A A+ +P +
Sbjct 169 ---------QGYAMPGSMAVASDSHSNTYGGVGCLGTPIVRTDAAAIWATGQTWWQIPPI 219
Query 150 VGVHL 154
V+L
Sbjct 220 ARVNL 224
> YGL009c
Length=779
Score = 34.7 bits (78), Expect = 0.080, Method: Composition-based stats.
Identities = 35/127 (27%), Positives = 56/127 (44%), Gaps = 25/127 (19%)
Query 25 VDVDL-VVDHSVQVDYSRSESALQRNLE--------KEMERNKERFAFLKWGSKAFNNMR 75
VD L VDH++ + ++ +L ++ K +E N ++F +G ++ R
Sbjct 61 VDCTLATVDHNIPTESRKNFKSLDTFIKQTDSRLQVKTLENNVKQFGVPYFG---MSDAR 117
Query 76 IVPPGSGIVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNGLGVLGWGVGGIEAEA 135
GIVH + E +G L +VV DSHT+ G L +G+G E E
Sbjct 118 -----QGIVHTIGPE--------EGFTLPGTTVVCGDSHTSTHGAFGSLAFGIGTSEVEH 164
Query 136 VMLGQHI 142
V+ Q I
Sbjct 165 VLATQTI 171
> SPAC9E9.03
Length=758
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 32/120 (26%), Positives = 48/120 (40%), Gaps = 24/120 (20%)
Query 31 VDHSVQVDYSRSESAL--------QRNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSG 82
VDH++ D + + R +E N + F +G N+ R G
Sbjct 67 VDHNIPTDPRKDMKDIASFIHQPDSRTQVLALENNIKEFGLTYYG---MNDRR-----QG 118
Query 83 IVHQVNLEFLASVTMDKGGLLYPDSVVGTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHI 142
IVH + E +G L ++V DSHT+ G L +G+G E E V+ Q I
Sbjct 119 IVHVIGPE--------QGFTLPGTTLVCGDSHTSTHGAFGALAFGIGTSEVEHVLATQTI 170
> At1g56570
Length=611
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 87 VNLEFLASVTMDKGGLLYPDSVVGTD-SHTTMVNGLGVLGWGVGGIEAEAVML 138
+N+ SVTM+ L++ D V D + TT++ G LG G+GG++ ML
Sbjct 153 MNMYATCSVTMEAACLIFRDIKVKNDVTWTTLITGFTHLGDGIGGLKMYKQML 205
> Hs4557779
Length=504
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 2/68 (2%)
Query 28 DLVVDHSVQVDYSRSESALQRNLEKEMERNKERFAFLKWGS--KAFNNMRIVPPGSGIVH 85
+L+ D SV + + NL + +E + + A L+ G + + R VPPGS V
Sbjct 144 NLLRDKSVLEEEKKRLRQENENLARRLESSSQEVARLRRGQCPQTRDTARAVPPGSREVS 203
Query 86 QVNLEFLA 93
NL+ LA
Sbjct 204 TWNLDTLA 211
> At4g24490
Length=683
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 10/72 (13%)
Query 6 AMRDAMARLGGDPRLINPLVDVDLVVDHSVQVDYSRSESALQRNLEKEMERNKERFAFLK 65
A+ D +AR+ DP L++ ++D +L V ESAL++N + + ++ K
Sbjct 68 AVEDRLARIEPDPNLVSAILDEELRV----------VESALRQNFKSYGAWHHRKWVLSK 117
Query 66 WGSKAFNNMRIV 77
S N +R++
Sbjct 118 GHSSVGNELRLL 129
> At4g13430
Length=509
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 25/101 (24%), Positives = 40/101 (39%), Gaps = 10/101 (9%)
Query 48 RNLEKEMERNKERFAFLKWGSKAFNNMRIVPPGSGIVHQVNLEFLASVTMDKGGLLYPDS 107
RN++ E +E+ + N + P G+ H V + + G P
Sbjct 143 RNVDIMREHCREQNIKYFYDITDLGNFKANPDYKGVCH---------VALAQEGHCRPGE 193
Query 108 VV-GTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISMVLP 147
V+ GTDSHT G G+G +A V+ I + +P
Sbjct 194 VLLGTDSHTCTAGAFGQFATGIGNTDAGFVLGTGKILLKVP 234
> SPBC3H7.06c
Length=467
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 19/35 (54%), Gaps = 1/35 (2%)
Query 110 GTDSHTTMVNGLGVLGWGVGGIEAEAVMLGQHISM 144
G DSH+ V G+G L W G E+EA + SM
Sbjct 205 GWDSHSAYVPGIGFLVWHTGR-ESEAQLFQPEASM 238
Lambda K H
0.319 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40