bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5322_orf1
Length=90
Score E
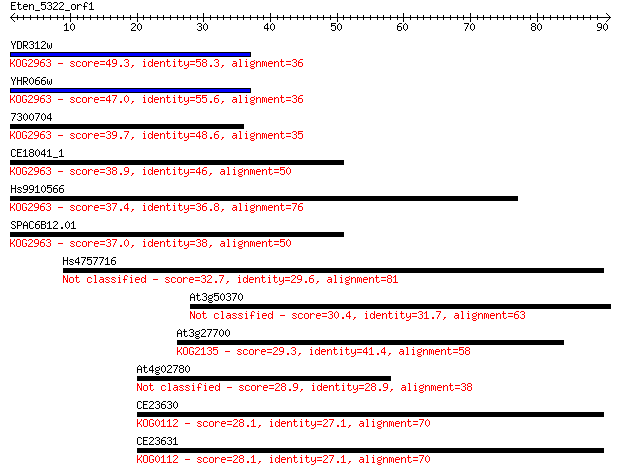
Sequences producing significant alignments: (Bits) Value
YDR312w 49.3 2e-06
YHR066w 47.0 9e-06
7300704 39.7 0.001
CE18041_1 38.9 0.002
Hs9910566 37.4 0.006
SPAC6B12.01 37.0 0.009
Hs4757716 32.7 0.15
At3g50370 30.4 0.78
At3g27700 29.3 1.9
At4g02780 28.9 2.3
CE23630 28.1 4.2
CE23631 28.1 4.3
> YDR312w
Length=453
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 31/36 (86%), Gaps = 0/36 (0%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKALKKK 36
L LKL KIED +C+GKVL+H+ + K++EE+KAL+K+
Sbjct 335 LTLKLVKIEDGICSGKVLHHEFVQKSSEEIKALEKR 370
> YHR066w
Length=453
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 20/36 (55%), Positives = 31/36 (86%), Gaps = 0/36 (0%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKALKKK 36
L LKL KIE+ +C+GKVL+H+ + K++EE+KAL+K+
Sbjct 335 LTLKLVKIEEGICSGKVLHHEFVQKSSEEIKALEKR 370
> 7300704
Length=460
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKALKK 35
L ++L KIE+ + TG+VLYH H++KT +E + L+K
Sbjct 277 LTMQLIKIEEGLLTGEVLYHDHVVKTEDEKETLRK 311
> CE18041_1
Length=400
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKALK----KKEQLLRAKREEERQK 50
L L+L KIE+ + G+VLYHKH KT +EL L+ KK+Q+ + + +E Q+
Sbjct 347 LTLELVKIEEGIDEGEVLYHKHNAKTPDELIKLRAHMDKKKQMKKRREQESEQR 400
> Hs9910566
Length=473
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 47/84 (55%), Gaps = 8/84 (9%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKA-LKKKEQLLRAKREEERQKLLELLER-- 57
+ L+L K+++ V GKV++H + KT EEL+A L+ KE+ LR K + + Q+ + +
Sbjct 279 MTLQLIKVQEGVGEGKVMFHSFVSKTEEELQAILEAKEKKLRLKAQRQAQQAQNVQRKQE 338
Query 58 -----RDKTAAAAKQAAAAAADEE 76
R K+ K+A +DEE
Sbjct 339 QREAHRKKSLEGMKKARVGGSDEE 362
> SPAC6B12.01
Length=361
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 1 LELKLFKIEDDVCTGKVLYHKHILKTAEELKALKKKEQLLRAKREEERQK 50
+ L+L KI +D GKVLYH H+ K+ EE+K + RA +E+ +++
Sbjct 268 MTLELIKITEDAMGGKVLYHSHVHKSKEEIKQQDNFHEQSRALKEKRKKE 317
> Hs4757716
Length=293
Score = 32.7 bits (73), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 43/81 (53%), Gaps = 1/81 (1%)
Query 9 EDDVCTGKVLYHKHILKTAEELKALKKKEQLLRAKREEERQKLLELLERRDKTAAAAKQA 68
EDD C +++ K + EEL + ++ E+ K EE+R KL EL +R+++ AA
Sbjct 97 EDDDCRYVMIFKKEFAPSDEELDSYRRGEEWDPQKAEEKR-KLKELAQRQEEEAAQQGPV 155
Query 69 AAAAADEEQQSSSKKRGRGSS 89
+ A + + S G+G++
Sbjct 156 VVSPASDYKDKYSHLIGKGAA 176
> At3g50370
Length=2152
Score = 30.4 bits (67), Expect = 0.78, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Query 28 EELKALKKKEQLLRAKREEERQKLLELLERRDKTAAAAKQAAAAAADEEQQSSSKKRGRG 87
E L+A ++ E+L ++K EE+ + +E E R K AA K +Q+ + K
Sbjct 533 ERLEATRRAEELRKSKEEEKHRLFME--EERRKQAAKQKLLELEEKISRRQAEAAKGCSS 590
Query 88 SST 90
SST
Sbjct 591 SST 593
> At3g27700
Length=831
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 16/74 (21%)
Query 26 TAEELK-ALKKKEQLLRAKREEERQKLLELLE---------------RRDKTAAAAKQAA 69
T E LK L+KK+++L KR E R+KL L + +R K A+ A
Sbjct 594 TLERLKETLRKKQEMLEQKRNEYRKKLATLEKQGTVVKREEADEPDAKRVKLDTASDSGA 653
Query 70 AAAADEEQQSSSKK 83
A A+ + + S+ KK
Sbjct 654 AIASPKTESSTDKK 667
> At4g02780
Length=802
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 11/38 (28%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 20 HKHILKTAEELKALKKKEQLLRAKREEERQKLLELLER 57
H H ++ AE + + Q L+A+R +E++K ++ +E+
Sbjct 713 HTHFVRLAEIINRICLPRQYLKARRNDEKEKTIKSMEK 750
> CE23630
Length=2526
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 0/70 (0%)
Query 20 HKHILKTAEELKALKKKEQLLRAKREEERQKLLELLERRDKTAAAAKQAAAAAADEEQQS 79
H+ L E + +K + + ER+ + + + R D AAAK +A D+++Q
Sbjct 1244 HQQRLTEDRENRKRQKSLTAYSSDEQGERKNVPKRMRRDDSEDAAAKHPGWSAKDDQKQR 1303
Query 80 SSKKRGRGSS 89
K R SS
Sbjct 1304 KRKLEHRRSS 1313
> CE23631
Length=2722
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 0/70 (0%)
Query 20 HKHILKTAEELKALKKKEQLLRAKREEERQKLLELLERRDKTAAAAKQAAAAAADEEQQS 79
H+ L E + +K + + ER+ + + + R D AAAK +A D+++Q
Sbjct 1244 HQQRLTEDRENRKRQKSLTAYSSDEQGERKNVPKRMRRDDSEDAAAKHPGWSAKDDQKQR 1303
Query 80 SSKKRGRGSS 89
K R SS
Sbjct 1304 KRKLEHRRSS 1313
Lambda K H
0.308 0.123 0.308
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177758614
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40