bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
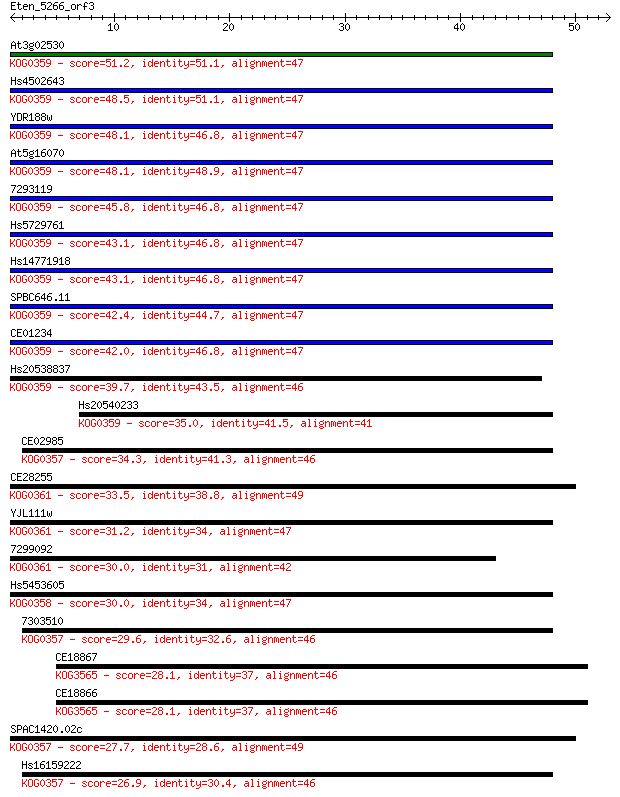
Query= Eten_5266_orf3
Length=52
Score E
Sequences producing significant alignments: (Bits) Value
At3g02530 51.2 4e-07
Hs4502643 48.5 3e-06
YDR188w 48.1 4e-06
At5g16070 48.1 4e-06
7293119 45.8 2e-05
Hs5729761 43.1 1e-04
Hs14771918 43.1 1e-04
SPBC646.11 42.4 2e-04
CE01234 42.0 3e-04
Hs20538837 39.7 0.001
Hs20540233 35.0 0.032
CE02985 34.3 0.061
CE28255 33.5 0.092
YJL111w 31.2 0.54
7299092 30.0 1.0
Hs5453605 30.0 1.2
7303510 29.6 1.3
CE18867 28.1 4.4
CE18866 28.1 4.5
SPAC1420.02c 27.7 5.7
Hs16159222 26.9 9.6
> At3g02530
Length=535
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR+L GF+ A++ TLQ LD K V + PD+E+L VART
Sbjct 111 EGMHPRVLVDGFEIAKRATLQFLDTFKTPVVMGDEPDKEILKMVART 157
> Hs4502643
Length=531
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 33/47 (70%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR++ GF+ A++K LQ L+EVKV + DRE L+ VART
Sbjct 112 EGLHPRIITEGFEAAKEKALQFLEEVKVSREM----DRETLIDVART 154
> YDR188w
Length=546
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 32/47 (68%), Gaps = 1/47 (2%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EGVHPR++ GF+ AR+++++ LDE K+ DRE LL VAR+
Sbjct 111 EGVHPRIITDGFEIARKESMKFLDEFKIS-KTNLSNDREFLLQVARS 156
> At5g16070
Length=540
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR+L GF+ A++ TLQ LD K V + D+E+L VART
Sbjct 116 EGMHPRVLVDGFEIAKRATLQFLDNFKTPVVMGDEVDKEILKMVART 162
> 7293119
Length=533
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 34/47 (72%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR++ GF+KAR K L++LD+VKV V + +++ L+ VA T
Sbjct 112 EGLHPRIMTDGFEKARDKALEVLDQVKVPVEI----NKKNLVEVANT 154
> Hs5729761
Length=540
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 34/47 (72%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR++ GF+ A+ K L++L+EVKV ++ R++LL VART
Sbjct 111 EGLHPRIIAEGFEAAKIKALEVLEEVKVTKEMK----RKILLDVART 153
> Hs14771918
Length=530
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 34/47 (72%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR++ GF+ A+ K L++L+EVKV ++ R++LL VART
Sbjct 112 EGLHPRIIAEGFEAAKIKALEVLEEVKVTKEMK----RKILLDVART 154
> SPBC646.11
Length=535
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HP L+ GF+ A+ + L LD K V DRE+LL+VA+T
Sbjct 110 EGLHPSLISDGFNLAKNEALTFLDSFKTDFEV----DREVLLNVAKT 152
> CE01234
Length=539
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+HPR++ GF+ A KTL+LL++ K + VE R+LL+ V RT
Sbjct 112 EGLHPRIVTEGFEWANTKTLELLEKFKKEAPVE----RDLLVEVCRT 154
> Hs20538837
Length=190
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 4/46 (8%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVAR 46
EG+HPR++ GF+ ++K L L+EVKV + D+E L VAR
Sbjct 110 EGLHPRIITEGFEAVKEKALHFLEEVKVSREM----DKETLKDVAR 151
> Hs20540233
Length=323
Score = 35.0 bits (79), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 28/41 (68%), Gaps = 4/41 (9%)
Query 7 LLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
++ GF+ A++K L+ L+EVK++ DRE L++VART
Sbjct 63 IITEGFEAAKEKALRFLEEVKIR----KEMDRETLINVART 99
> CE02985
Length=542
Score = 34.3 bits (77), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 2 GVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
G+HP + GFD A +K L+ LD + K VE +RE L+ A+T
Sbjct 128 GIHPIKIADGFDLACKKALETLDSISDKFPVE---NRERLVETAQT 170
> CE28255
Length=535
Score = 33.5 bits (75), Expect = 0.092, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 29/49 (59%), Gaps = 0/49 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVARTLS 49
+GVHP+LL KA +KTL+ L ++++K+ E L+ A TLS
Sbjct 114 DGVHPQLLIRAIGKACEKTLKNLADLEIKINGETELREMLVKCAATTLS 162
> YJL111w
Length=550
Score = 31.2 bits (69), Expect = 0.54, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
EG+ L+ G+ KA ++ ++E+ V + E RELL ART
Sbjct 118 EGISSHLIMKGYRKAVSLAVEKINELAVDITSEKSSGRELLERCART 164
> 7299092
Length=501
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLL 42
EGVHPR++ KA Q ++ ++E+ V++ + + + LL
Sbjct 70 EGVHPRVIIKAIRKALQLCMEKINEMAVQIVEQSKDQQRALL 111
> Hs5453605
Length=539
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
+G+HP ++ F KA +K +++L ++ V + DRE LL+ A T
Sbjct 126 KGIHPTIISESFQKALEKGIEILTDMSRPVELS---DRETLLNSATT 169
> 7303510
Length=542
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 2 GVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
G+HP + GF+ A Q ++ LD + V+P+ ++E L+ +A T
Sbjct 128 GIHPIRIADGFELAAQCAIKQLDAIAQPFPVDPK-NKEPLIQIAMT 172
> CE18867
Length=785
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 5 PRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVARTLSV 50
P++L + F A +K+L D VKV+ AV E S A T+S+
Sbjct 698 PKILRSSFSGAIRKSLSTPDSVKVETAVTVTALFEFAKSSAETMSI 743
> CE18866
Length=783
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 5 PRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVARTLSV 50
P++L + F A +K+L D VKV+ AV E S A T+S+
Sbjct 698 PKILRSSFSGAIRKSLSTPDSVKVETAVTVTALFEFAKSSAETMSI 743
> SPAC1420.02c
Length=546
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 1 EGVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVARTLS 49
+G+HP + G++KA Q ++ LD + V P L S +L
Sbjct 128 KGIHPIRIADGYEKACQVAVKHLDAISDVVDFSPENTTNLFRSAKTSLG 176
> Hs16159222
Length=541
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 2 GVHPRLLCAGFDKARQKTLQLLDEVKVKVAVEPRPDRELLLSVART 47
G+HP + G+++A + ++ LD++ V V+ + D E L+ A+T
Sbjct 127 GIHPIRIADGYEQAARVAIEHLDKISDSVLVDIK-DTEPLIQTAKT 171
Lambda K H
0.320 0.137 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1158678716
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40