bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5236_orf1
Length=90
Score E
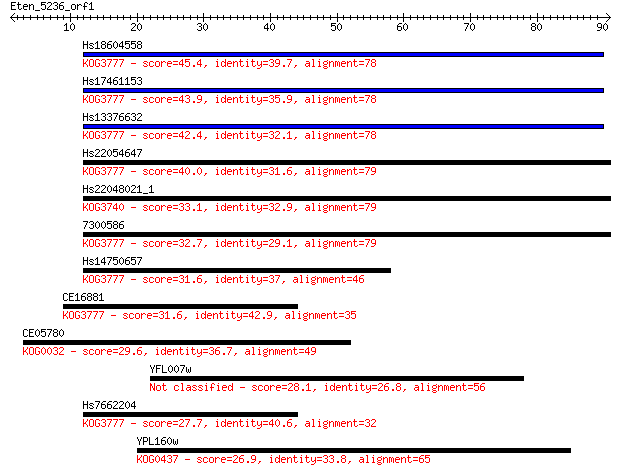
Sequences producing significant alignments: (Bits) Value
Hs18604558 45.4 2e-05
Hs17461153 43.9 6e-05
Hs13376632 42.4 2e-04
Hs22054647 40.0 0.001
Hs22048021_1 33.1 0.12
7300586 32.7 0.18
Hs14750657 31.6 0.35
CE16881 31.6 0.41
CE05780 29.6 1.5
YFL007w 28.1 4.3
Hs7662204 27.7 4.8
YPL160w 26.9 9.2
> Hs18604558
Length=650
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 47/83 (56%), Gaps = 24/83 (28%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQERAQMPVSYTRMPD 70
G ++FS +GI A+D++ ERG+K + VFVP A R+E+++ PD
Sbjct 27 GNKEVFSCRGIKLAVDWFLERGHKDITVFVP-----------AWRKEQSR--------PD 67
Query 71 CI----EVLRKLHREGILICTPS 89
+ E+LRKL +E IL+ TPS
Sbjct 68 ALITDQEILRKLEKEKILVFTPS 90
> Hs17461153
Length=849
Score = 43.9 bits (102), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 45/86 (52%), Gaps = 17/86 (19%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQERAQMP-------V 63
G + FS +GI A+D++ ++G+K + VFVP + + Q R P V
Sbjct 205 GNKEEFSCRGIQLAVDWFLDKGHKDITVFVPAW---------RKEQSRPDAPITDDFCSV 255
Query 64 SYTRMPDCIEVLRKLHREGILICTPS 89
+Y + ++LRKL +E IL+ TPS
Sbjct 256 AYNHILTDQDILRKLEKEKILVFTPS 281
> Hs13376632
Length=599
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 44/79 (55%), Gaps = 16/79 (20%)
Query 12 GPDKLFSTKGILAALDYYRERGY-KVRVFVPEYVLSYNSYSDARRQERAQMPVSYTRMPD 70
G ++FS +GIL A++++ ERG+ + VFVP + + Q R +P++
Sbjct 150 GNKEVFSCRGILLAVNWFLERGHTDITVFVPSW---------RKEQPRPDVPITDQH--- 197
Query 71 CIEVLRKLHREGILICTPS 89
+LR+L ++ IL+ TPS
Sbjct 198 ---ILRELEKKKILVFTPS 213
> Hs22054647
Length=824
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 42/80 (52%), Gaps = 16/80 (20%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQERAQMPVSYTRMPD 70
G + FS +GI A+D++R+RG+ ++VFVP + + RA P+
Sbjct 280 GNKETFSCRGIKLAVDWFRDRGHTYIKVFVPSW---------RKDPPRADTPIREQ---- 326
Query 71 CIEVLRKLHREGILICTPSQ 90
VL +L R+ +L+ TPS+
Sbjct 327 --HVLAELERQAVLVYTPSR 344
> Hs22048021_1
Length=945
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 35/80 (43%), Gaps = 19/80 (23%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQERAQMPVSYTRMPD 70
G FS +GI A+ ++ RG++ V VFVP + L N RR +
Sbjct 689 GLQHFFSCRGIAMAVQFFWNRGHREVTVFVPTWQLKKN-----RRVRESHF--------- 734
Query 71 CIEVLRKLHREGILICTPSQ 90
L KLH +L TPSQ
Sbjct 735 ----LTKLHSLKMLSITPSQ 750
> 7300586
Length=536
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 41/80 (51%), Gaps = 18/80 (22%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQERAQMPVSYTRMPD 70
G + +FS +GI +D++R+RG++ + FVP + R+E A ++
Sbjct 143 GNNLVFSCRGIRICVDWFRQRGHRDITAFVPNW-----------RKEMANNNIADQ---- 187
Query 71 CIEVLRKLHREGILICTPSQ 90
E+L +L E L+ TPS+
Sbjct 188 --ELLYELEHERYLVFTPSR 205
> Hs14750657
Length=719
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 29/47 (61%), Gaps = 5/47 (10%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEYVLSYNSYSDARRQE 57
G FS++GI A+ Y+ +RG++ + VFVP++ S DA+ +E
Sbjct 493 GLQHYFSSRGIAIAVQYFWDRGHRDITVFVPQWRFS----KDAKVRE 535
> CE16881
Length=369
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 25/36 (69%), Gaps = 4/36 (11%)
Query 9 GRKGPDKLFSTKGILAALDYYRERGY-KVRVFVPEY 43
GRK ++FS G+ L+Y+ ERG+ +V +F+P+Y
Sbjct 274 GRK---EVFSCAGLRECLNYFLERGHPEVLIFIPQY 306
> CE05780
Length=336
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 3 ETVRGTGR-KGPDKLFSTKGILAALDYYRERGYKVRVFVPEYVLSYNSYS 51
ET+R GR + PD T + +AL+Y ER R PE +L + +S
Sbjct 159 ETIRKNGRIEEPDAAIITLQVASALNYLHERNVVHRDVKPENLLLVDKFS 208
> YFL007w
Length=1804
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 15/59 (25%), Positives = 32/59 (54%), Gaps = 3/59 (5%)
Query 22 ILAALDYYRERGYKVRVFVPEYVLSYNSYSDARRQERAQMPV---SYTRMPDCIEVLRK 77
+L LD ++ G+K+R+F+ +V S +++ +RA + T+ P+ I ++ K
Sbjct 1446 LLDKLDNEKDMGWKIRMFILRFVTQIQSNLESKPDKRAVFSIISQISTKHPEIIHLVVK 1504
> Hs7662204
Length=896
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query 12 GPDKLFSTKGILAALDYYRERGYK-VRVFVPEY 43
G K FS +GI A++Y+ + G + + VFVP++
Sbjct 632 GLKKFFSCRGIAIAVEYFWKLGNRNITVFVPQW 664
> YPL160w
Length=1090
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 8/72 (11%)
Query 20 KGILAALDYYRERGYKVRVFVPEYVLSYNSYS---DARRQERAQMPVSYTRMPD----CI 72
KG+LAALDY R +R + + S DA + + + +S + P+ C+
Sbjct 906 KGVLAALDYLRNLQRSIREGEGQALKKKKGKSAEIDASKPVKLTLLISES-FPEWQSQCV 964
Query 73 EVLRKLHREGIL 84
E++RKL E L
Sbjct 965 EIVRKLFSEQTL 976
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177758614
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40