bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5122_orf1
Length=79
Score E
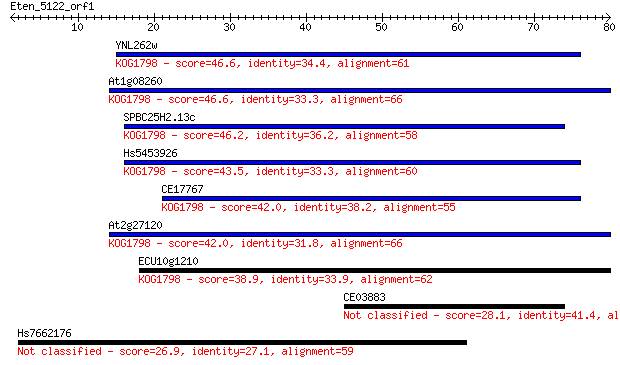
Sequences producing significant alignments: (Bits) Value
YNL262w 46.6 1e-05
At1g08260 46.6 1e-05
SPBC25H2.13c 46.2 1e-05
Hs5453926 43.5 9e-05
CE17767 42.0 3e-04
At2g27120 42.0 3e-04
ECU10g1210 38.9 0.003
CE03883 28.1 4.5
Hs7662176 26.9 9.6
> YNL262w
Length=2222
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 15 RKLRFEFPTWLLNFAVYRRFRNEMFLSF-CPERKSFLKEIKNEILFELDGPWKGMFLPAS 73
+KL +P +LN+ V+++F N + P + +N I FE+DGP+K M LP+S
Sbjct 899 KKLYLSYPCSMLNYRVHQKFTNHQYQELKDPLNYIYETHSENTIFFEVDGPYKAMILPSS 958
Query 74 EK 75
++
Sbjct 959 KE 960
> At1g08260
Length=2271
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 14 VRKLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPA 72
++KL +P +LN V + N+ + + P RK++ + I FE+DGP+K M +PA
Sbjct 855 MKKLTISYPCVMLNVDVAKNNTNDQYQTLVDPVRKTYKSHSECSIEFEVDGPYKAMIIPA 914
Query 73 SEKSDSL 79
S++ L
Sbjct 915 SKEEGIL 921
> SPBC25H2.13c
Length=2199
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSF-CPERKSFLKEIKNEILFELDGPWKGMFLPAS 73
K+ +P +LN V+ +F N + + PE+ + +N I FE+DGP++ M LPAS
Sbjct 885 KVFISYPCVMLNHLVHEKFTNHQYSALKDPEKLVYETTSENSIFFEVDGPYRAMILPAS 943
> Hs5453926
Length=2286
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 1/61 (1%)
Query 16 KLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPASE 74
K+ +P +LN V F N+ + P +++ +N I FE+DGP+ M LPAS+
Sbjct 887 KVTISYPGAMLNIMVKEGFTNDQYQELAEPSSLTYVTRSENSIFFEVDGPYLAMILPASK 946
Query 75 K 75
+
Sbjct 947 E 947
> CE17767
Length=2144
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 21 FPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASEK 75
+P +LN VY F N + + + S+ K +N I FE+DGP++ M LPAS++
Sbjct 872 YPGAMLNALVYEGFTNHQYHTL-EKDGSYSKSSENSIYFEVDGPYQCMILPASKE 925
> At2g27120
Length=2154
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 14 VRKLRFEFPTWLLNFAVYRRFRNEMFLSFC-PERKSFLKEIKNEILFELDGPWKGMFLPA 72
++K +P +LN V + N+ + + P RK++ + I FE+DGP+K M +PA
Sbjct 848 MKKFTISYPCVILNVDVAKNNSNDQYQTLVDPVRKTYNSRSECSIEFEVDGPYKAMIIPA 907
Query 73 SEKSDSL 79
S++ L
Sbjct 908 SKEEGIL 914
> ECU10g1210
Length=1799
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 18 RFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFELDGPWKGMFLPASEKSD 77
RF F + LLN V + F N + + ++ +N I FE+DGP++ M LP+S +
Sbjct 791 RFSFLSSLLNHFVCKEFTNWQYQERVDGK--YVMRAENSIFFEIDGPYRAMLLPSSTEES 848
Query 78 SL 79
L
Sbjct 849 KL 850
> CE03883
Length=272
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 45 ERKSFLKEIKNEILFELDGPWKGMFLPAS 73
ERK+ L +I N ++F WKG F+ S
Sbjct 13 ERKTTLIKISNSVIFPCSSDWKGAFIRLS 41
> Hs7662176
Length=673
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 25/59 (42%), Gaps = 0/59 (0%)
Query 2 ISSSSSSSSSRGVRKLRFEFPTWLLNFAVYRRFRNEMFLSFCPERKSFLKEIKNEILFE 60
+ + + G R LR E A RR R SF ER+ +L+E + I ++
Sbjct 558 VDGEAEAGGESGTRALRREVGRLQAELAAERRARERQGASFAEERRVWLEEKEKVIEYQ 616
Lambda K H
0.322 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1168763848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40