bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
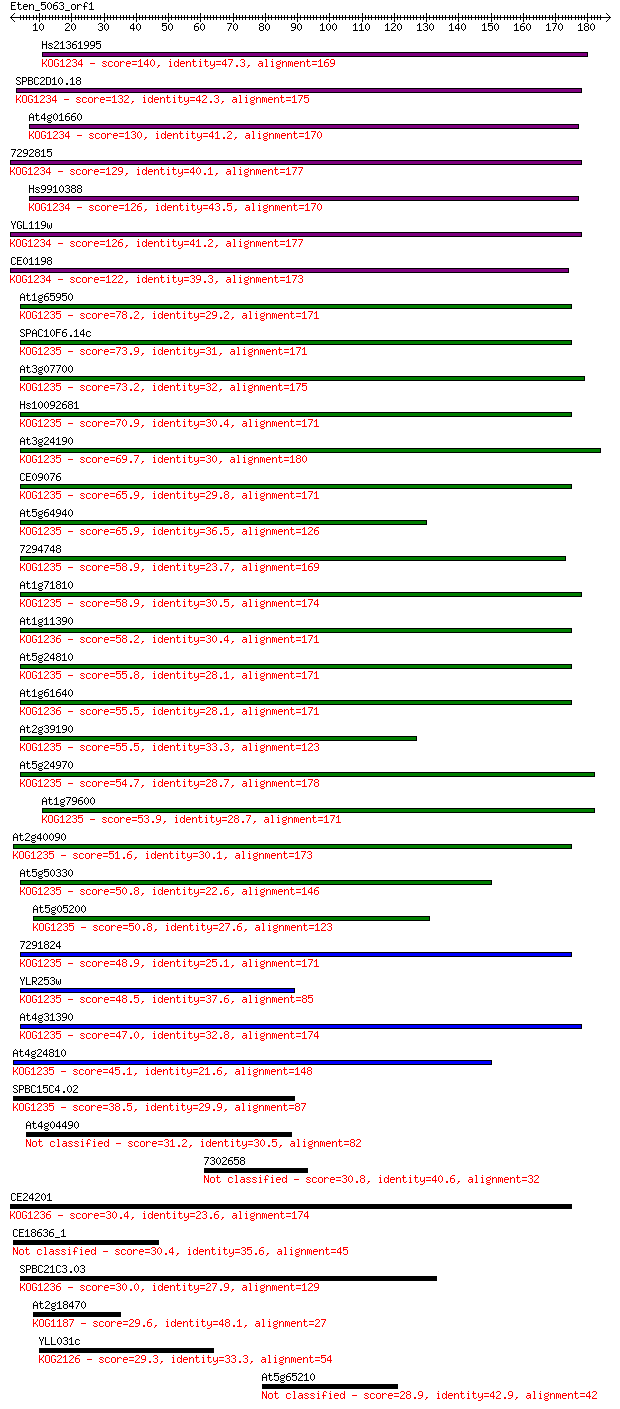
Query= Eten_5063_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
Hs21361995 140 1e-33
SPBC2D10.18 132 3e-31
At4g01660 130 2e-30
7292815 129 3e-30
Hs9910388 126 2e-29
YGL119w 126 3e-29
CE01198 122 4e-28
At1g65950 78.2 8e-15
SPAC10F6.14c 73.9 2e-13
At3g07700 73.2 3e-13
Hs10092681 70.9 1e-12
At3g24190 69.7 3e-12
CE09076 65.9 5e-11
At5g64940 65.9 5e-11
7294748 58.9 6e-09
At1g71810 58.9 6e-09
At1g11390 58.2 8e-09
At5g24810 55.8 5e-08
At1g61640 55.5 6e-08
At2g39190 55.5 7e-08
At5g24970 54.7 1e-07
At1g79600 53.9 2e-07
At2g40090 51.6 8e-07
At5g50330 50.8 2e-06
At5g05200 50.8 2e-06
7291824 48.9 5e-06
YLR253w 48.5 7e-06
At4g31390 47.0 2e-05
At4g24810 45.1 8e-05
SPBC15C4.02 38.5 0.008
At4g04490 31.2 1.4
7302658 30.8 1.8
CE24201 30.4 1.9
CE18636_1 30.4 2.3
SPBC21C3.03 30.0 2.8
At2g18470 29.6 3.2
YLL031c 29.3 4.2
At5g65210 28.9 7.0
> Hs21361995
Length=386
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 80/169 (47%), Positives = 106/169 (62%), Gaps = 3/169 (1%)
Query 11 PFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPSSFFLDRL 70
PFAAASIGQVH+ +L+ G EVAVKIQ+PGIA SI SDV+NLL +L++S LP+ F ++
Sbjct 56 PFAAASIGQVHQGLLRDGTEVAVKIQYPGIAQSIQSDVQNLLAVLKMSAALPAGLFAEQS 115
Query 71 SQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVITTQFVRAVP 130
Q LQQEL EC+Y EA F LL D R+P + + L T RV+ + VP
Sbjct 116 LQALQQELAWECDYRREAACAQNFRQLLAND--PFFRVPAVVKELCTTRVLGMELAGGVP 173
Query 131 FAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDGDA 179
Q SQ+ RN++ LL L LRE+F +R + TD N ANF++D +
Sbjct 174 LD-QCQGLSQDLRNQICFQLLTLCLRELFEFRFMQTDPNWANFLYDASS 221
> SPBC2D10.18
Length=610
Score = 132 bits (333), Expect = 3e-31, Method: Composition-based stats.
Identities = 74/176 (42%), Positives = 105/176 (59%), Gaps = 5/176 (2%)
Query 3 HFLEFDMNPFAAASIGQVHRAVLKTGE-EVAVKIQFPGIAASITSDVKNLLLLLQLSLLL 61
H+ EFD P AAASIGQVHRA L + EV VK+Q+PG+ +SI SD+ NL LL+ S +L
Sbjct 293 HYSEFDRKPMAAASIGQVHRARLASNHMEVVVKVQYPGVMSSIDSDLNNLAYLLKASRIL 352
Query 62 PSSFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVI 121
P FL+ ++EL EC+Y EA F F LL+ D ++P ++R + VI
Sbjct 353 PKGLFLENSLAAARKELKWECDYEREAAFAERFGSLLKND--SDFKVPMVFREASGPSVI 410
Query 122 TTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDG 177
T +++ + A+ Q SQ RN +G LL + LREI Y + TD N +NF+++G
Sbjct 411 TLEYLHGI--ALGKQKYSQATRNHIGYLLTKQCLREISEYHFMQTDPNWSNFLYNG 464
> At4g01660
Length=623
Score = 130 bits (326), Expect = 2e-30, Method: Composition-based stats.
Identities = 70/170 (41%), Positives = 105/170 (61%), Gaps = 3/170 (1%)
Query 7 FDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPSSFF 66
FD P AAASIGQVHRAV K G EVA+KIQ+PG+A SI SD++N+ LL + L+P F
Sbjct 296 FDYEPLAAASIGQVHRAVTKDGLEVAMKIQYPGVANSIESDIENVRRLLNYTNLIPKGLF 355
Query 67 LDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVITTQFV 126
LDR + ++EL EC+Y EA F LL D P +P + ++++++TT+ +
Sbjct 356 LDRAIKVAKEELAQECDYEIEAVSQKRFRDLLS-DTP-GFYVPLVVDETSSKKILTTELI 413
Query 127 RAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWD 176
+P A Q+ R+ +G +L L L+E+F++R + TD N NF+++
Sbjct 414 SGIPIDKVA-LLDQKTRDYVGRKMLELTLKELFVFRFMQTDPNWGNFLYN 462
> 7292815
Length=999
Score = 129 bits (325), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 71/177 (40%), Positives = 106/177 (59%), Gaps = 3/177 (1%)
Query 1 RQHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLL 60
RQ F+ PFAAASIGQVHRA L G +VA+KIQ+PG+A SI SD+ NL+ +L++ +
Sbjct 343 RQRLKSFEDKPFAAASIGQVHRATLSDGMDVAIKIQYPGVAQSIESDIDNLVGMLKVWDV 402
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRV 120
P FF+D + + ++EL E +Y EA + F ++ +P + +P++ R LTT V
Sbjct 403 FPQGFFIDNVVRVAKRELQWEVDYDREAEYTEKFREMIA-PYPEYY-VPRVVRDLTTSSV 460
Query 121 ITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDG 177
+TT+ V VP S E R + +L+L LRE+F + TD N +NF++D
Sbjct 461 LTTELVPGVPLDKCFDL-SYEHRRHIAASVLKLCLRELFEIECMQTDPNWSNFLYDA 516
> Hs9910388
Length=368
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/170 (43%), Positives = 103/170 (60%), Gaps = 3/170 (1%)
Query 7 FDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPSSFF 66
F+ PFAAASIGQVH A +K G EVA+KIQ+PG+A SI SDV NL+ +L +S +LP F
Sbjct 52 FEERPFAAASIGQVHLARMKGGREVAMKIQYPGVAQSINSDVNNLMAVLNMSNMLPEGLF 111
Query 67 LDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVITTQFV 126
+ L L++EL EC+Y EA F LL + P +P++ L + V+TT+ V
Sbjct 112 PEHLIDVLRRELALECDYQREAACARKFRDLL-KGHP-FFYVPEIVDELCSPHVLTTELV 169
Query 127 RAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWD 176
P A+ SQE RN++ +L L LRE+F + + TD N +NF +D
Sbjct 170 SGFPLD-QAEGLSQEIRNEICYNILVLCLRELFEFHFMQTDPNWSNFFYD 218
> YGL119w
Length=501
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 73/177 (41%), Positives = 102/177 (57%), Gaps = 3/177 (1%)
Query 1 RQHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLL 60
+ F +FD P AAASIGQVH A L +G+ V VKIQ+PG+ SI SD+ +LL+LL S L
Sbjct 183 KTKFSKFDKIPMAAASIGQVHAAELPSGQRVVVKIQYPGVKESIDSDLNSLLMLLTASSL 242
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRV 120
LP FLD+ + EL EC+Y EA F LL+ D P +P ++ TT +
Sbjct 243 LPKGLFLDKTIANARTELKWECDYNREARALQKFEALLKDD-P-AFEVPHVFPEYTTDNI 300
Query 121 ITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDG 177
IT + + ASQE +N + E ++RL L EI ++ + TD N ANF+++G
Sbjct 301 ITMTRMEGTEI-MKLPKASQETKNFISENIMRLCLEEIATFKYMQTDPNWANFLYNG 356
> CE01198
Length=733
Score = 122 bits (306), Expect = 4e-28, Method: Composition-based stats.
Identities = 68/173 (39%), Positives = 101/173 (58%), Gaps = 3/173 (1%)
Query 1 RQHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLL 60
R+ F FD PFA ASIGQVH+AVLK G VAVK+Q+PG+A I SD+ NL+ +L + +
Sbjct 402 REKFEHFDDKPFACASIGQVHKAVLKDGRNVAVKVQYPGVAEGIDSDIDNLVSVLSVGGI 461
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRV 120
P FLD ++EL EC+Y EA F L+ D+ + + +P++ L++ RV
Sbjct 462 FPKGMFLDAFVGVARRELKQECDYEREARAMKKFRELIA-DW-QDVYVPEVIDELSSSRV 519
Query 121 ITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANF 173
+TT+ V P + Q R+ + + L L+EIF++R + TD N +NF
Sbjct 520 LTTELVYGKPVDACVE-EPQVVRDYIAGKFIELCLKEIFLWRFMQTDPNWSNF 571
> At1g65950
Length=505
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 50/174 (28%), Positives = 89/174 (51%), Gaps = 7/174 (4%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
+L FD P AAASI QVH AVLK +EVAVK+Q+PG+ ++ D + L + +
Sbjct 165 YLSFDEEPIAAASIAQVHHAVLKNHQEVAVKVQYPGLKQNMMLDTMIMSFLSKSVAKIFP 224
Query 64 SFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFP--RHLRLPQLYRHLTTQRVI 121
+ D L + + + E ++L EA S + ++F + + +P ++ TT +V+
Sbjct 225 EYRFDWLVYEFVKSISQELDFLQEAK----NSERIAKNFKHNKMITIPTVFSEFTTTQVL 280
Query 122 TTQFVRAVPF-AVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFM 174
T QF + V + + K+ ++L+ + IF++ I+ D +P N +
Sbjct 281 TMQFCKGFKVDDVESLKRTNVSPEKVAKVLVEVFAEMIFVHGFIHGDPHPGNIL 334
> SPAC10F6.14c
Length=535
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/180 (29%), Positives = 98/180 (54%), Gaps = 15/180 (8%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDV---KNLLLLLQLSLL 60
F + A+ASI QVHRAVL +GE+VAVKIQ P +A ++ D+ K ++ + +
Sbjct 177 FASIEKRAAASASIAQVHRAVLPSGEKVAVKIQKPDVAKQMSWDLLVYKYMMYVYDKWIF 236
Query 61 -LPSSFFLDRLSQQLQQELLSECNYLNEA-NFHLLFSFLLQRDFPR-HLRLPQLYRHLTT 117
+P F +D +S++L+ SE ++ EA N + + D+ R + +P++Y+ ++
Sbjct 237 HIPLYFTVDYVSERLR----SEVDFTTEANNSEHAREGVEETDYLRDKIYIPKVYKEISG 292
Query 118 QRVITTQFVRAVPFAVAAQTASQEDRNKMGEL---LLRLVLREIFIYRLINTDANPANFM 174
+RV+ T++ +P + QTA E E+ L R + ++F + ++ D +P N +
Sbjct 293 KRVMVTEWADGIP--LYDQTALSEAGMSKKEILTNLFRFLAFQMFHSKQVHCDPHPGNIL 350
> At3g07700
Length=670
Score = 73.2 bits (178), Expect = 3e-13, Method: Composition-based stats.
Identities = 56/181 (30%), Positives = 90/181 (49%), Gaps = 11/181 (6%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
+ EF+ P AAAS+GQVHRAVL GE+V VK+Q PG+ D++NL L+ +
Sbjct 257 YKEFEEQPIAAASLGQVHRAVLHNGEKVVVKVQRPGLKKLFDIDLRNLKLIAEY-FQKSE 315
Query 64 SFFLDR---LSQQLQQELLSECNYLNEANFHLLFSFLLQRDFP--RHLRLPQLYRHLTTQ 118
SF + + ++ L E +Y+NEA F +RDF +R+P +Y +
Sbjct 316 SFGTNDWVGIYEECALILYQEIDYINEAKNADRF----RRDFRNINWVRVPLVYWDYSAM 371
Query 119 RVITTQFVRAVPF-AVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDG 177
+V+T ++V V + A A +R+++ + L +I + D +P N D
Sbjct 372 KVLTLEYVPGVKINNLDALAARGFNRSRIASRAIEAYLIQILKTGFFHADPHPGNLAIDV 431
Query 178 D 178
D
Sbjct 432 D 432
> Hs10092681
Length=455
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 52/174 (29%), Positives = 88/174 (50%), Gaps = 7/174 (4%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNL-LLLLQLSLLLP 62
F FD P AS+ QVH+AVL G VAVK+Q P + A + D+ + +L+L + L P
Sbjct 78 FQSFDDTPLGTASLAQVHKAVLHDGRTVAVKVQHPKVRAQSSKDILLMEVLVLAVKQLFP 137
Query 63 SSFFLDRLSQQLQQELLSECNYLNEA-NFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVI 121
F+ L + ++ L E ++LNE N + L DF L++P+++ L+T+RV+
Sbjct 138 EFEFM-WLVDEAKKNLPLELDFLNEGRNAEKVSQMLRHFDF---LKVPRIHWDLSTERVL 193
Query 122 TTQFVRAVPFAVAAQTASQE-DRNKMGELLLRLVLREIFIYRLINTDANPANFM 174
+FV + D N++ L ++ IF+ ++ D +P N +
Sbjct 194 LMEFVDGGQVNDRDYMERNKIDVNEISRHLGKMYSEMIFVNGFVHCDPHPGNVL 247
> At3g24190
Length=809
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/195 (27%), Positives = 97/195 (49%), Gaps = 23/195 (11%)
Query 4 FLEFDMNPFAAASIGQVHRAVLK-TGEEVAVKIQFPGIAASITSD---VKNLLLLL---- 55
+ E +P AAAS+GQV++ LK G+ VAVK+Q P + ++T D ++NL L L
Sbjct 259 YSELSPSPIAAASLGQVYKGRLKENGDLVAVKVQRPFVLETVTVDLFVIRNLGLFLRKFP 318
Query 56 QLSLLLPSSF--FLD--RLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQL 111
Q L + FLD L + E +Y+NE F+ ++++D P + +P+
Sbjct 319 QARFCLTAQLMQFLDVVGLVDEWAARFFEELDYVNEGENGTYFAEMMKKDLP-QVVVPKT 377
Query 112 YRHLTTQRVITTQFVRAVPFAVAAQTASQEDRNKMGELL---LRLVLREIFIYRLINTDA 168
Y+ T+++V+TT ++ + SQ + +GEL+ + L+++ + D
Sbjct 378 YQKYTSRKVLTTSWID-------GEKLSQSIESDVGELVNVGVICYLKQLLDTGFFHADP 430
Query 169 NPANFMWDGDAPLPV 183
+P N + D L +
Sbjct 431 HPGNMIRTPDGKLAI 445
> CE09076
Length=512
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 51/182 (28%), Positives = 89/182 (48%), Gaps = 22/182 (12%)
Query 4 FLEFDMNPFAAASIGQVHRAVLK-TGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLP 62
F EF P AAS+ QVH+A LK +GE VAVK+Q + + +DV + L++++ +
Sbjct 152 FSEFSEKPVGAASLAQVHKAKLKESGETVAVKVQHKRVYKNSRTDVNTMEFLVKVADAVF 211
Query 63 SSFFLDRLSQQLQQELLSECNYLNEAN---------FHLLFSFLLQRDFPRHLRLPQLYR 113
F L L ++++ L +E ++L+EA HL F LR+P++
Sbjct 212 PEFRLMWLVDEIKKNLPNELDFLHEAKNADEAAQRFKHLKF-----------LRIPKIKY 260
Query 114 HLTTQRVITTQFVRAVPF-AVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPAN 172
LTT RV+T +F V + D + + + + + IF+ +++D +P N
Sbjct 261 DLTTTRVLTMEFCEGAHVDDVEYLKKNNIDPHDVCMKIGKTISEMIFLQGYLHSDPHPGN 320
Query 173 FM 174
+
Sbjct 321 VL 322
> At5g64940
Length=795
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 46/131 (35%), Positives = 71/131 (54%), Gaps = 8/131 (6%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQ-LSLLLP 62
F FD P AAAS+GQVHRA LK G+EV +K+Q PG+ D+KNL ++ + L + P
Sbjct 286 FDRFDYEPIAAASLGQVHRARLK-GQEVVLKVQRPGLKDLFDIDLKNLRVIAEYLQKVDP 344
Query 63 SSFFLDR----LSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQ 118
S R + + L E +Y EA LF+ +D ++++P +Y TT
Sbjct 345 KSDGAKRDWVAIYDECASVLYQEIDYTKEAANSELFANNF-KDL-EYVKVPSIYWEYTTP 402
Query 119 RVITTQFVRAV 129
+V+T ++V +
Sbjct 403 QVLTMEYVPGI 413
> 7294748
Length=557
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 40/174 (22%), Positives = 86/174 (49%), Gaps = 11/174 (6%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
+ EFD P AAAS+ QV +A L +GE+VAVK+Q+ + SD+ ++ L +
Sbjct 172 YQEFDYQPVAAASLAQVFKARLPSGEQVAVKVQYNDLQKRFISDLGTIIFLQDIVEFFFK 231
Query 64 SFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPR--HLRLPQLYRHLTTQRVI 121
+ + L++ L+ E N+L E + +D + ++ +P+++ T RV+
Sbjct 232 DYNFGWILNDLRKNLVLELNFLQEGQ----NAERCAKDMEKFSYVHVPKVHWSYTKTRVL 287
Query 122 TTQFVRAVPFAVAAQTASQEDRNKMGELLLRL---VLREIFIYRLINTDANPAN 172
T +++ ++ ++++ + ++ ++L +IF ++ D +P N
Sbjct 288 TLEWMDGC--KISDLKTIEKEKLSLKDIDVKLFEAFAEQIFYTGFVHADPHPGN 339
> At1g71810
Length=671
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 53/188 (28%), Positives = 92/188 (48%), Gaps = 25/188 (13%)
Query 4 FLEFDMNPFAAASIGQVHRAVL-KTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLP 62
F E P AAAS+GQV++A L ++G+ VAVK+Q PG+ A+I D L+L L+
Sbjct 187 FSEISPEPVAAASLGQVYQARLRRSGKVVAVKVQRPGVRAAIALD--TLILRYIAGLIKK 244
Query 63 SSFFLDRLSQ----------QLQQELLS-ECNYLNEANFHLLFSFLLQRDFPRHLRLPQL 111
+ F L +++ LLS E +YLNEA + F L + + +P++
Sbjct 245 AGRFNSDLEAVVDEWATSLFKVKDTLLSQEMDYLNEAQNGIKFRKLYGGI--KDVLVPKM 302
Query 112 YRHLTTQRVITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPA 171
Y +T +V+ ++V Q ++ + + E+ + ++ Y + D +P
Sbjct 303 YTEYSTSKVLVMEWVE-------GQKLNEVNDLYLVEVGVYCSFNQLLEYGFYHADPHPG 355
Query 172 NFM--WDG 177
NF+ +DG
Sbjct 356 NFLRTYDG 363
> At1g11390
Length=456
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 52/187 (27%), Positives = 96/187 (51%), Gaps = 22/187 (11%)
Query 4 FLEFDMNPFAAASIGQVHRAVLK---TGEE-----VAVKIQFPGIAASITSDVKNLLLLL 55
F EFD P A+ SI QVHRA L+ G++ VAVK++ PG+ SI D + L+
Sbjct 84 FEEFDEVPVASGSIAQVHRASLRFQYPGQKSKSSLVAVKVRHPGVGESIRRDFVIINLVA 143
Query 56 QLSLLLPSSFFLDRLSQQLQQ---ELLSECNYLNEANFHL---LFSFLLQRD--FPRHLR 107
++S L+P+ +L RL + +QQ +LS+ + EA+ HL +++F +D FP+ +
Sbjct 144 KISTLIPALKWL-RLDESVQQFGVFMLSQVDLAREAS-HLSRFIYNFRRWKDVSFPKPV- 200
Query 108 LPQLYRHLTTQRVITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTD 167
P ++ + + T + +V V + + ++ + +L+ + + I+ D
Sbjct 201 YPLVHPAVLVE---TYEHGESVARYVDGMEGHEWIKTRLAHIGTHALLKMLLVDNFIHAD 257
Query 168 ANPANFM 174
+P N +
Sbjct 258 MHPGNIL 264
> At5g24810
Length=962
Score = 55.8 bits (133), Expect = 5e-08, Method: Composition-based stats.
Identities = 48/181 (26%), Positives = 83/181 (45%), Gaps = 12/181 (6%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
F +F P A ASI QVHRA L G++V VK+Q GI A I D+KN ++
Sbjct 118 FTDFVDEPLATASIAQVHRATLANGQDVVVKVQHDGIRAIILEDLKNAKSIVDWIAWAEP 177
Query 64 SFFLDRLSQQLQQELLSECNYLNEA-NFHLLFSFLLQRDFPRHLR--------LPQLYRH 114
+ + + + +E E ++ EA N + L + +R +P + +
Sbjct 178 QYNFNPMIDEWCKEAPRELDFNIEAENTRTVSGNLGCKKTNDEVRSANRVDVLIPDIIQ- 236
Query 115 LTTQRVITTQFVRAVPFA-VAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANF 173
+++ V+ +++ V V + A D+ K+ E + R +IF+ N D +P NF
Sbjct 237 -SSESVLILEYMDGVRLNDVESLDAFGVDKQKIVEEITRAYAHQIFVDGFFNGDPHPGNF 295
Query 174 M 174
+
Sbjct 296 L 296
> At1g61640
Length=428
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 48/186 (25%), Positives = 88/186 (47%), Gaps = 20/186 (10%)
Query 4 FLEFDMNPFAAASIGQVHRAVLK--------TGEEVAVKIQFPGIAASITSDVKNLLLLL 55
F EFD P A+ SI QVHRA LK EVAVK++ P + ++ D + +
Sbjct 84 FEEFDEAPVASGSIAQVHRASLKFQYAGQKVKSSEVAVKVRHPCVEETMKRDFVIINFVA 143
Query 56 QLSLLLPSSFF--LDRLSQQLQQELLSECNYLNEANFHL---LFSFLLQRD--FPRHLRL 108
+L+ +P + LD QQ +LS+ + EA+ HL +++F +D FP+ +
Sbjct 144 RLTTFIPGLNWLRLDECVQQFSVYMLSQVDLSREAS-HLSRFIYNFRGWKDVSFPKPI-Y 201
Query 109 PQLYRHLTTQRVITTQFVRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDA 168
P ++ + + T + +V V ++ + K+ + +L+ + + I+ D
Sbjct 202 PLIHPAVLVE---TYEHGESVARYVDGSEGQEKLKAKVAHIGTNALLKMLLVDNFIHADM 258
Query 169 NPANFM 174
+P N +
Sbjct 259 HPGNIL 264
> At2g39190
Length=791
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 65/133 (48%), Gaps = 21/133 (15%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
F +F AAAS GQV+R G +VAVK+Q P + ++ D+ +L L L +L
Sbjct 266 FSQFSQETVAAASFGQVYRGRTLDGADVAVKVQRPDLRHAVLRDI--YILRLGLGVLRKV 323
Query 64 S-------FFLDRLSQQLQQELLSECNYLNEANF---HLLFSFLLQRDFPRHLRLPQLYR 113
+ + D L L EL N + F H FS+ +R+P++Y+
Sbjct 324 AKRENDIRVYADELGMGLAGELDFTLEAANASEFQEAHARFSY---------IRVPKVYQ 374
Query 114 HLTTQRVITTQFV 126
HLT +RV+T +++
Sbjct 375 HLTRKRVLTMEWM 387
> At5g24970
Length=715
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 51/202 (25%), Positives = 95/202 (47%), Gaps = 27/202 (13%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLL---QLSLL 60
F + + P AAAS+GQV++A L +G+ VAVK+Q PG++ +T D LL + QL
Sbjct 197 FADISLKPVAAASLGQVYKAHLHSGQLVAVKVQRPGMSLILTRDA--LLFKMIGGQLKRF 254
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEAN----FHLLFSF------LLQRDFPRHL---- 106
+ L ++ + + E +Y+ EA F L+SF + PR++
Sbjct 255 AKARKDLLVAVNEMVRHMFDEIDYVLEAKNAERFASLYSFDSGNEQIDDNAGPRNMSRNH 314
Query 107 -----RLPQLYRHLTTQRVITTQFVRAVPFA--VAAQTASQEDRNKMGELLLRLVLREIF 159
++P++Y + T V+T +++ + + + AS DR + + L L+++
Sbjct 315 RAENIKVPKIYWNFTRTAVLTMEWIDGIKLTDEIKLKRASL-DRRDLIDQGLSCSLKQLL 373
Query 160 IYRLINTDANPANFMWDGDAPL 181
+ D +P N + + L
Sbjct 374 EVGFFHADPHPGNLVATKEGSL 395
> At1g79600
Length=711
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 49/182 (26%), Positives = 88/182 (48%), Gaps = 20/182 (10%)
Query 11 PFAAASIGQVHRAVLK-TGEEVAVKIQFPGIAASITSD---VKNLLLLLQLSLLLPSS-- 64
P AAAS+GQV++A L+ +G+ VAVK+Q PGI +I D ++ + L+ + ++
Sbjct 221 PIAAASLGQVYKAQLRYSGQVVAVKVQRPGIEEAIGLDFYLIRGVGKLINKYVDFITTDV 280
Query 65 -FFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVITT 123
+D + ++ QEL NY+ EA F L + +P ++ T+++V+T
Sbjct 281 LTLIDEFACRVYQEL----NYVQEAQNARRFKKLYADK--ADVLVPDIFWDYTSRKVLTM 334
Query 124 QFVRAVP----FAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFMWDGDA 179
++V A+ +Q D G ++ LR++ Y + D +P N + D
Sbjct 335 EWVEGTKLNEQLAIESQGLKVLDLVNTG---IQCSLRQLLEYGFFHADPHPGNLLATPDG 391
Query 180 PL 181
L
Sbjct 392 KL 393
> At2g40090
Length=544
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 52/187 (27%), Positives = 92/187 (49%), Gaps = 24/187 (12%)
Query 2 QHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQ-LSLL 60
Q F EFD P A+AS+ QVH A G++VAVK+Q + + +D + +L+ L +
Sbjct 161 QVFAEFDPVPIASASLAQVHVARTHDGKKVAVKVQHAHMTDTAAADTAAVGVLVNTLHRI 220
Query 61 LPS---SFFLDRLSQQLQQELLSECNYLNEA--NFHLLFSF-LLQRDFPRHLRLPQLYRH 114
PS + LD +S+ L +EL ++L EA N L +F L ++ P +Y +
Sbjct 221 FPSFDYRWLLDEMSESLPKEL----DFLVEAKNNEKCLDNFRKLSPHIAEYVYAPTIYWN 276
Query 115 LTTQRVITTQFVRAVPFAVAAQTASQEDRNKMG-------ELLLRLVLREIFIYRLINTD 167
L+T +++T +F+ AQ + K+G +L+ + +F + ++ D
Sbjct 277 LSTSKLLTMEFMD------GAQVNDVDKIRKLGIQPYEVSKLVSQTFAEMMFKHGFVHCD 330
Query 168 ANPANFM 174
+ AN +
Sbjct 331 PHAANLI 337
> At5g50330
Length=423
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/147 (22%), Positives = 67/147 (45%), Gaps = 16/147 (10%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
F FD P +ASI QV Q PGI + +D++NL L
Sbjct 131 FETFDEKPLGSASIAQV---------------QHPGIERLMMTDIRNLQLFALYMQRTDI 175
Query 64 SFFLDRLSQQLQQELLSECNYLNEAN-FHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVIT 122
F L +++++++++ E ++ EAN + FL + + + +P++ R + T+RV+
Sbjct 176 KFDLHSITKEMEKQIGYEFDFKREANAMERIRCFLYENNKKSPVLVPRVLRDMVTKRVLV 235
Query 123 TQFVRAVPFAVAAQTASQEDRNKMGEL 149
+++ +P ++ N G++
Sbjct 236 MEYINGIPILSIGDEMAKRGINPHGKI 262
> At5g05200
Length=509
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 67/128 (52%), Gaps = 5/128 (3%)
Query 8 DMNPFAAASIGQVHRAVLK-TGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPSSF- 65
D P A+ASI QVH A L+ + E+V +K+ PGI + +D+ + ++ ++ L F
Sbjct 166 DPTPIASASIAQVHGARLRGSQEDVVIKVLKPGIEDFLVADLNFIYVVSRIFEFLSPEFS 225
Query 66 --FLDRLSQQLQQELLSECNYLNEA-NFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVIT 122
L + + +++ +L E ++ EA N +L P++Y++ +++RV+T
Sbjct 226 RTSLVGIVKDIRESMLEEVDFNKEAQNIESFKRYLETMGLTGQATAPRVYKYCSSRRVLT 285
Query 123 TQFVRAVP 130
+ + VP
Sbjct 286 MERLYGVP 293
> 7291824
Length=501
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/178 (24%), Positives = 80/178 (44%), Gaps = 32/178 (17%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
F F+ P AS+ QVH+A LKTGE VA + A I D
Sbjct 147 FDSFEREPLGTASLAQVHKARLKTGELVATMELAVNVLARIFPD---------------- 190
Query 64 SFFLDRLSQQLQQELLSECNYLNEANFHLLFSFLLQRDFPRH--LRLPQLYRHLTTQRVI 121
F + L ++ ++ L E ++LNE + + + F ++ LR+P++Y ++ RV+
Sbjct 191 -FKIHWLVEESKKNLPIELDFLNEGR----NAEKVAKQFKKYSWLRVPKIYWKYSSSRVL 245
Query 122 TTQF-----VRAVPFAVAAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFM 174
++ V + + + S N++G+L ++ R F++ +D +P N +
Sbjct 246 VMEYLEGGHVTDLDYIRRNKIDSFAVANRIGQLYSEMIFRTGFVH----SDPHPGNIL 299
> YLR253w
Length=569
Score = 48.5 bits (114), Expect = 7e-06, Method: Composition-based stats.
Identities = 32/89 (35%), Positives = 42/89 (47%), Gaps = 4/89 (4%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKT----GEEVAVKIQFPGIAASITSDVKNLLLLLQLSL 59
FLEF+ P AS+ QVH A LK G VAVK Q P + I DV + +L
Sbjct 176 FLEFNKTPIGVASLAQVHVAKLKNSDGKGSSVAVKCQHPSLKEFIPLDVMLTRTVFELLD 235
Query 60 LLPSSFFLDRLSQQLQQELLSECNYLNEA 88
+ + L L +LQ + E N+ EA
Sbjct 236 VFFPDYPLTWLGDELQSSIYVELNFTKEA 264
> At4g31390
Length=596
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 57/204 (27%), Positives = 92/204 (45%), Gaps = 40/204 (19%)
Query 4 FLEFDMNPFAAASIGQVHRAVLK-TGEEVAVK---------------IQFPGIAASITSD 47
F + AAAS+GQV+RA L+ TGE+VA+K +Q P I I D
Sbjct 220 FSKISSQTIAAASLGQVYRATLRATGEDVAIKVLLNENFIVATEFIQVQRPQIEPIIYRD 279
Query 48 VKNLLLLLQLSLLLPSSFFLDRLS-------QQLQQELLSECNYLNEANFHLLFSFLLQ- 99
L L L+ L + F L +L + ++LL E +Y EA + FL
Sbjct 280 ---LFLFRTLASFL-NGFSLQKLGCNAELIVDEFGEKLLEELDYTLEA--RNIEDFLENF 333
Query 100 RDFPRHLRLPQLYRHLTTQRVITTQFVRAV----PFAVAAQTASQEDRNKMGELLLRLVL 155
+D P +++P +Y++L RV+ +++ + P A+ D N + + L
Sbjct 334 KDDP-TVKIPGVYKNLCGPRVLVMEWIDGIRCTDPQAIKDAGI---DLNGFLTVGVSAAL 389
Query 156 REIFIYRLINTDANPANF--MWDG 177
R++ + L + D +P N M DG
Sbjct 390 RQLLEFGLFHGDPHPGNIFAMQDG 413
> At4g24810
Length=410
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/149 (21%), Positives = 67/149 (44%), Gaps = 16/149 (10%)
Query 2 QHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLL 61
Q F FD P +ASI QV Q PG+ + D++NL +
Sbjct 129 QVFETFDEKPLGSASIAQV---------------QHPGVEKLMMVDIRNLQIFALYMQKT 173
Query 62 PSSFFLDRLSQQLQQELLSECNYLNEAN-FHLLFSFLLQRDFPRHLRLPQLYRHLTTQRV 120
F L +++++++++ E ++ EAN + FL + + +P+++ +L T++V
Sbjct 174 DIKFDLFSMTKEIEKQIGYEFDFKREANAMEKIRRFLYDNNRKSPVLVPRVFPNLVTRKV 233
Query 121 ITTQFVRAVPFAVAAQTASQEDRNKMGEL 149
+ +F+ +P ++ N G++
Sbjct 234 LVMEFMNGIPILSLGDEMAKRGINPHGKM 262
> SPBC15C4.02
Length=594
Score = 38.5 bits (88), Expect = 0.008, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 44/88 (50%), Gaps = 1/88 (1%)
Query 2 QHFLEFDMNPFAAASIGQVHRAVLKTGEE-VAVKIQFPGIAASITSDVKNLLLLLQLSLL 60
+ F EFD AS+ QVH+A LK + VAVK+Q P ++ + D+ + +
Sbjct 213 ETFDEFDPIALGVASLAQVHKARLKDSDVWVAVKVQHPSVSLNSPLDLSMTRWVFKAIKT 272
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEA 88
F L L+ ++++ L E ++ EA
Sbjct 273 FFPDFKLMWLADEIERSLPQELDFTREA 300
> At4g04490
Length=658
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 14/83 (16%)
Query 6 EFDM-NPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPSS 64
EF + N G V++ +L +G+E+AVK + G + + KN +LLL
Sbjct 339 EFSLENKLGQGGFGSVYKGILPSGQEIAVK-RLAGGSGQGELEFKNEVLLLT-------- 389
Query 65 FFLDRLSQQLQQELLSECNYLNE 87
RL + +LL CN NE
Sbjct 390 ----RLQHRNLVKLLGFCNEGNE 408
> 7302658
Length=546
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 61 LPSSFFLDRLSQQLQQELLSECNYLNEANFHL 92
+PS F DR Q+LQ C++LN N L
Sbjct 49 IPSEIFRDRTQQELQTITCKRCHFLNHYNIAL 80
> CE24201
Length=600
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 41/221 (18%), Positives = 86/221 (38%), Gaps = 47/221 (21%)
Query 1 RQHFLEFDMNPFAAASIGQVHRAVLKTGE-EVAVKIQFPGIAASITSDV------KNLLL 53
+Q FLE + + I QV+R + E E A ++P + T + K++
Sbjct 191 KQVFLEIEPFSIGSGCIAQVYRGTVDISELEKATGKEYPELKGRTTQRIAIKVADKDVDK 250
Query 54 LLQLSL------------LLPSSFFLDRLS--QQLQQELLSECNYLNEANFHLLFS---- 95
++L L ++P+ ++LD +Q + L + + NEA FS
Sbjct 251 QIELDLSILRSGAWLMQHIVPALWYLDPTGALEQFEMVLRRQVDLRNEAKALKKFSDNFS 310
Query 96 -------FLLQRDFPRHLRLPQLYRHLTTQRVITTQ---------------FVRAVPFAV 133
F + + +++ + + R++ + F++ + F +
Sbjct 311 SKETGIKFPIVLGYTKNVIVETFEEGIYINRLVAEEGQPEIFENETFFRYLFLKIIDFEM 370
Query 134 AAQTASQEDRNKMGELLLRLVLREIFIYRLINTDANPANFM 174
SQ R ++ L R +L+ IF+ ++ D +P N +
Sbjct 371 LTARQSQAVRRRIALLGARALLKMIFVDNFVHGDLHPGNIL 411
> CE18636_1
Length=1096
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 2 QHFLEFDMNPFAAASIGQVHRAVLKTGEEVAVKIQFPGIAASITS 46
Q FL NPF IG++H A T E V V+ G++ S+++
Sbjct 49 QTFLWRQTNPFLGDKIGKLHEASCVTFERVVVQNILHGLSPSLSN 93
> SPBC21C3.03
Length=674
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 36/168 (21%), Positives = 64/168 (38%), Gaps = 49/168 (29%)
Query 4 FLEFDMNPFAAASIGQVHRAVLKTG-----------------------------EEVAVK 34
F+ F+ P +I QV+ A +K ++VA+K
Sbjct 245 FVNFNPIPIGVGAIAQVYTATIKKAATQQDNSYFTSMLQSIGFRQKNISDAPVTQDVAIK 304
Query 35 IQFPGIAASITSDVKNLLLLLQLSLLLPSSFFLD----------RLSQQLQQELLSECNY 84
+ P + I+ D++ L +L L+PS +L L+QQL L E +
Sbjct 305 VLHPNVEKYISLDLQILGFFAKLINLVPSMKWLSLPDEVKVFGAMLNQQLN--LHYEALH 362
Query 85 LNEANFHLLFSFLLQRDFPRHLRLPQLYRHLTTQRVITTQFVRAVPFA 132
LN+ F L R++ P Y TT +++ ++ +P +
Sbjct 363 LNQ--------FRLNFRGNRYVEFPAPYDDYTTNQILLEDYMPGIPLS 402
> At2g18470
Length=633
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 8 DMNPFAAASIGQVHRAVLKTGEEVAVK 34
D N G VH+ VL +G+EVAVK
Sbjct 286 DANLLGQGGFGYVHKGVLPSGKEVAVK 312
> YLL031c
Length=1017
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 10 NPFAAASIGQVHRAV-LKTGEEVAVKIQFPGIAASITSDVKNLLLLLQLSLLLPS 63
+P +G ++ V L+T EE+ K + IA IT +L+LL+ ++ L+PS
Sbjct 420 DPEKFVKLGHKYQKVFLQTCEELWAKFDYYSIATGITLLATSLVLLISITKLIPS 474
> At5g65210
Length=368
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 5/47 (10%)
Query 79 LSECNYLNEAN-----FHLLFSFLLQRDFPRHLRLPQLYRHLTTQRV 120
L E +Y+ + N L SF+ Q D RH L Q+YR LTT++
Sbjct 293 LGEGSYIPQVNSAMDRLEALVSFVNQADHLRHETLQQMYRILTTRQA 339
Lambda K H
0.326 0.139 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40