bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5061_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
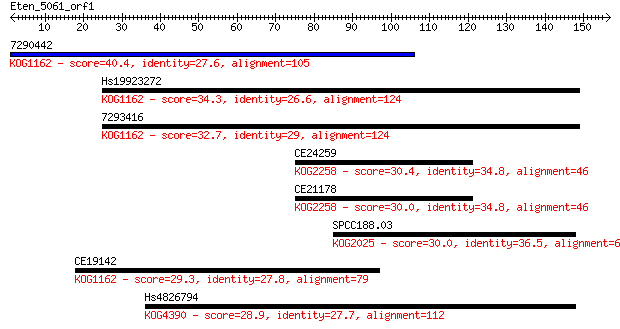
7290442 40.4 0.001
Hs19923272 34.3 0.090
7293416 32.7 0.31
CE24259 30.4 1.3
CE21178 30.0 1.7
SPCC188.03 30.0 2.2
CE19142 29.3 2.9
Hs4826794 28.9 4.7
> 7290442
Length=649
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 49/105 (46%), Gaps = 4/105 (3%)
Query 1 FRLGFAWSFIWLSTASAMAVMERYGVNYCFLLDLAPRGQTSSVSFFCTAALHFVLTTATC 60
FR F W A+ +A ++ GVN+ + ++ PR +F A +L +
Sbjct 263 FRGPFTWVIFNFYMAANVAGWQQAGVNHILIFEIDPRSHLQPATFLEIACTFGILWALSM 322
Query 61 ALYLADSKLHLLGDGQMYHFYPLGLLLLQLLLLLFPSRALTYKAR 105
+L + L+G Y F PLGL+L+ + LL+ P + + AR
Sbjct 323 LGFLYND---LIGVSDPYVF-PLGLILIMVGLLVVPLPIMNWPAR 363
> Hs19923272
Length=696
Score = 34.3 bits (77), Expect = 0.090, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 50/125 (40%), Gaps = 10/125 (8%)
Query 25 GVNYCFLLDLAPRGQTSSVSFFCTAALHFVL-TTATCALYLADSKLHLLGDGQMYHFYPL 83
GVN+ + +L PR S F A +L + A + A + + YPL
Sbjct 296 GVNHVLIFELNPRSNLSHQHLFEIAGFLGILWCLSLLACFFAPISV------IPTYVYPL 349
Query 84 GLLLLQLLLLLFPSRALTYKARRQVLGAFFTVLKAGAFTVRDVKLVANIIGDVLTSLAKP 143
L + L+ P++ YK+R +L F V A V + D L SL+
Sbjct 350 ALYGFMVFFLINPTKTFYYKSRFWLLKLLFRVFTA---PFHKVGFADFWLADQLNSLSVI 406
Query 144 LGDLQ 148
L DL+
Sbjct 407 LMDLE 411
> 7293416
Length=681
Score = 32.7 bits (73), Expect = 0.31, Method: Composition-based stats.
Identities = 36/132 (27%), Positives = 52/132 (39%), Gaps = 24/132 (18%)
Query 25 GVNYCFLLDLAPRGQTSSVSFFCTAALHFVLTTATCALYLADSKLHLLGDGQMYHFYPLG 84
GVN+ + +L PR S AA+ V+ T + +L + L + PL
Sbjct 285 GVNHVLIFELDPRNHLSEQHLMELAAIFGVIWTLSMLSFLYSASLAIPA-----FINPLT 339
Query 85 LLLLQLLLLLFPSRALTYKAR--------RQVLGAFFTVLKAGAFTVRDVKLVANIIGDV 136
L L+ +L L P L + AR R V FF V A + +GD
Sbjct 340 LTLIMVLFLANPFHVLYHDARFWLWRITGRCVSAPFFHVGFADFW-----------LGDQ 388
Query 137 LTSLAKPLGDLQ 148
L SLA + D +
Sbjct 389 LNSLATAILDFE 400
> CE24259
Length=298
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 75 GQMYHFYPLGLLLLQLLLLLFPSRALTYKARRQVLGAFFTVLKAGA 120
G +Y YP GLLL+ L L +F K + + FF+ K G
Sbjct 22 GAIYLIYPPGLLLIPLSLFIFSYTTKNEKCSSKNVDTFFSGFKIGG 67
> CE21178
Length=356
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 75 GQMYHFYPLGLLLLQLLLLLFPSRALTYKARRQVLGAFFTVLKAGA 120
G +Y YP GLLL+ L L +F K + + FF+ K G
Sbjct 22 GAIYLIYPPGLLLIPLSLFIFSYTTKNEKCSSKNVDTFFSGFKIGG 67
> SPCC188.03
Length=875
Score = 30.0 bits (66), Expect = 2.2, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 29/63 (46%), Gaps = 2/63 (3%)
Query 85 LLLLQLLLLLFPSRALTYKARRQVLGAFFTVLKAGAFTVRDVKLVANIIGDVLTSLAKPL 144
L L L++ F + A RQVLG FF V GA + +A I D L SL +
Sbjct 638 LFLKPLIIQYFEPNTVDNHALRQVLGYFFPVYAFGAH--ENQWRIATIFCDALLSLLEIY 695
Query 145 GDL 147
DL
Sbjct 696 RDL 698
> CE19142
Length=710
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 5/79 (6%)
Query 18 MAVMERYGVNYCFLLDLAPRGQTSSVSFFCTAALHFVLTTATCALYLADSKLHLLGDGQM 77
MA GVN+ + ++ PR S + A+ +L + YL LH+
Sbjct 281 MAGWAAAGVNHVLIFEVDPRNHLSYQTLMQIASFMIMLWSFAVLAYLYAHMLHIPPFAP- 339
Query 78 YHFYPLGLLLLQLLLLLFP 96
PL L+++ L+LLL P
Sbjct 340 ----PLALMVVCLILLLNP 354
> Hs4826794
Length=655
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 31/117 (26%), Positives = 46/117 (39%), Gaps = 14/117 (11%)
Query 36 PRGQTSSVSFFCTAALHFVLTTATCALYLADSKLHLLGDGQMYHFYPLGLLLLQLLLLLF 95
P G+ SV+FFC L TA ++ + L L Y F + ++ ++ ++
Sbjct 217 PCGERYSVAFFC-------LDTACVMIFTGEYLLRLFAAPSRYRFIRSVMSIIDVVAIMP 269
Query 96 PSRALTYKARRQVLGAF-----FTVLKAGAFTVRDVKLVANIIGDVLTSLAKPLGDL 147
L V GAF F V + F+ L I+G L S A LG L
Sbjct 270 YYIGLVMTNNEDVSGAFVTLRVFRVFRIFKFSRHSQGL--RILGYTLKSCASELGFL 324
Lambda K H
0.331 0.143 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40