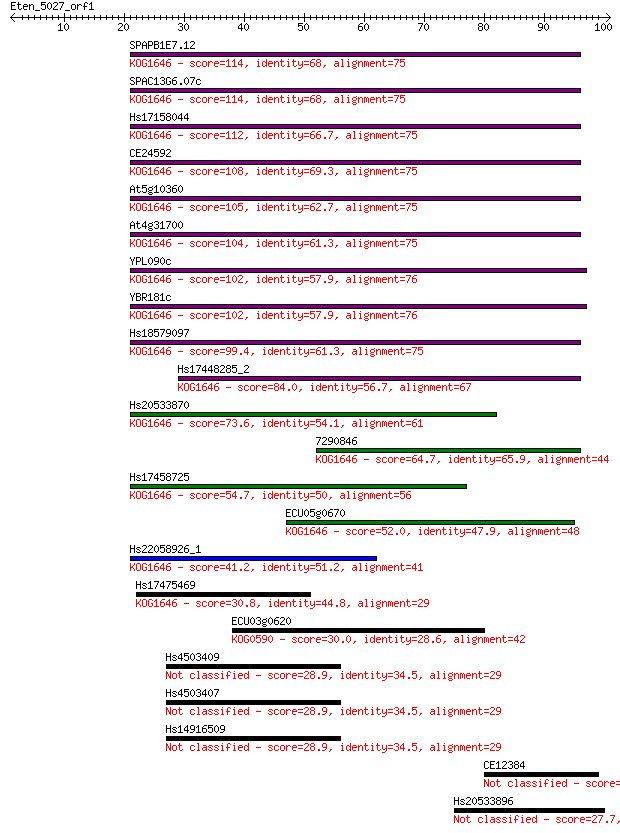
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5027_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
SPAPB1E7.12 114 4e-26
SPAC13G6.07c 114 4e-26
Hs17158044 112 2e-25
CE24592 108 2e-24
At5g10360 105 2e-23
At4g31700 104 4e-23
YPL090c 102 2e-22
YBR181c 102 2e-22
Hs18579097 99.4 2e-21
Hs17448285_2 84.0 6e-17
Hs20533870 73.6 9e-14
7290846 64.7 4e-11
Hs17458725 54.7 4e-08
ECU05g0670 52.0 3e-07
Hs22058926_1 41.2 4e-04
Hs17475469 30.8 0.70
ECU03g0620 30.0 1.1
Hs4503409 28.9 2.8
Hs4503407 28.9 2.8
Hs14916509 28.9 2.8
CE12384 28.5 3.0
Hs20533896 27.7 6.0
> SPAPB1E7.12
Length=239
Score = 114 bits (285), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 51/75 (68%), Positives = 62/75 (82%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ PA G QK +E+DD+++L F E+RMG EVPGDS+G EF GY+F+ITGGNDKQG
Sbjct 1 MKLNISYPANGTQKLIEIDDDRRLRVFMEKRMGQEVPGDSVGPEFAGYVFKITGGNDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L HRVRL
Sbjct 61 FPMFQGVLLPHRVRL 75
> SPAC13G6.07c
Length=239
Score = 114 bits (285), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 51/75 (68%), Positives = 62/75 (82%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ PA G QK +E+DD+++L F E+RMG EVPGDS+G EF GY+F+ITGGNDKQG
Sbjct 1 MKLNISYPANGTQKLIEIDDDRRLRVFMEKRMGQEVPGDSVGPEFAGYVFKITGGNDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L HRVRL
Sbjct 61 FPMFQGVLLPHRVRL 75
> Hs17158044
Length=249
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 50/75 (66%), Positives = 61/75 (81%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ PA G QK +EVDDE+KL F+E+RM EV D+LGEE+KGY+ RI+GGNDKQG
Sbjct 1 MKLNISFPATGCQKLIEVDDERKLRTFYEKRMATEVAADALGEEWKGYVVRISGGNDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L + RVRL
Sbjct 61 FPMKQGVLTHGRVRL 75
> CE24592
Length=246
Score = 108 bits (271), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 52/75 (69%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
M+LN A PA GLQK+ EVD+EKKL FFE+RM EV D+LG+E+KGY+ RI GGNDKQG
Sbjct 1 MRLNFAYPATGLQKSFEVDEEKKLRLFFEKRMSQEVAIDALGDEWKGYVVRIGGGNDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QGIL N RVRL
Sbjct 61 FPMKQGILTNGRVRL 75
> At5g10360
Length=249
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 47/75 (62%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MK N+ANP G QK +E+DD++KL FF++R+ EV GD+LGEEFKGY+F+I GG DKQG
Sbjct 1 MKFNVANPTTGCQKKLEIDDDQKLRAFFDKRLSQEVSGDALGEEFKGYVFKIMGGCDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L RVRL
Sbjct 61 FPMKQGVLTPGRVRL 75
> At4g31700
Length=250
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 46/75 (61%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MK N+ANP G QK +E+DD++KL F+++R+ EV GD+LGEEFKGY+F+I GG DKQG
Sbjct 1 MKFNVANPTTGCQKKLEIDDDQKLRAFYDKRISQEVSGDALGEEFKGYVFKIKGGCDKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L RVRL
Sbjct 61 FPMKQGVLTPGRVRL 75
> YPL090c
Length=236
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 44/76 (57%), Positives = 61/76 (80%), Gaps = 0/76 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ P G QKT E+DDE ++ FF++R+G EV G+++G+EFKGY+F+I+GGNDKQG
Sbjct 1 MKLNISYPVNGSQKTFEIDDEHRIRVFFDKRIGQEVDGEAVGDEFKGYVFKISGGNDKQG 60
Query 81 FPMMQGILANHRVRLC 96
FPM QG+L R++L
Sbjct 61 FPMKQGVLLPTRIKLL 76
> YBR181c
Length=236
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 44/76 (57%), Positives = 61/76 (80%), Gaps = 0/76 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ P G QKT E+DDE ++ FF++R+G EV G+++G+EFKGY+F+I+GGNDKQG
Sbjct 1 MKLNISYPVNGSQKTFEIDDEHRIRVFFDKRIGQEVDGEAVGDEFKGYVFKISGGNDKQG 60
Query 81 FPMMQGILANHRVRLC 96
FPM QG+L R++L
Sbjct 61 FPMKQGVLLPTRIKLL 76
> Hs18579097
Length=240
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/75 (61%), Positives = 57/75 (76%), Gaps = 0/75 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLNI+ P G QK +EVDDE KL F+E+ M EV D+LGEE+KGY+ RI+GGN+KQG
Sbjct 1 MKLNISFPVTGCQKLIEVDDECKLRTFYEKLMATEVAADTLGEEWKGYVVRISGGNNKQG 60
Query 81 FPMMQGILANHRVRL 95
FPM QG+L + RV L
Sbjct 61 FPMKQGVLTHGRVHL 75
> Hs17448285_2
Length=155
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 38/67 (56%), Positives = 50/67 (74%), Gaps = 0/67 (0%)
Query 29 ACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQGFPMMQGIL 88
A G QK +EVDD++KL F+E+ M EV D+L EE+KGY+ I+GGNDKQ FPM +G+L
Sbjct 1 ATGCQKLIEVDDKRKLCTFYEKHMATEVAADALVEEWKGYVVLISGGNDKQWFPMKRGVL 60
Query 89 ANHRVRL 95
+ RVRL
Sbjct 61 IHGRVRL 67
> Hs20533870
Length=177
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 46/61 (75%), Gaps = 0/61 (0%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQG 80
MKLN + A G QK ++V+DE+KL F+E+ M E+ +LGEE+KGY+ RI+GGN+KQG
Sbjct 1 MKLNSSFLAIGCQKLIKVEDERKLRTFYEKHMATEIAAYALGEEWKGYVVRISGGNNKQG 60
Query 81 F 81
F
Sbjct 61 F 61
> 7290846
Length=217
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/44 (65%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 52 MGAEVPGDSLGEEFKGYLFRITGGNDKQGFPMMQGILANHRVRL 95
MG V D LG+E+KGY RI GGNDKQGFPM QG+L + RVRL
Sbjct 1 MGQVVEADILGDEWKGYQLRIAGGNDKQGFPMKQGVLTHGRVRL 44
> Hs17458725
Length=248
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 28/56 (50%), Positives = 38/56 (67%), Gaps = 1/56 (1%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGN 76
MKLNI A QK +EVD+E KL F+E+ M EV D+LG+E+KGY+ +T G+
Sbjct 1 MKLNIF-LATACQKLIEVDNEHKLHTFYEKCMATEVAADALGKEWKGYVGVLTHGH 55
> ECU05g0670
Length=203
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 47 FFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQGFPMMQGILANHRVR 94
+++++G + G LGEEF+G + ITGG+D QGFPM+ G L RVR
Sbjct 12 LYDKKIGDQFDGGILGEEFEGTIMEITGGDDYQGFPMVSGHLTKKRVR 59
> Hs22058926_1
Length=53
Score = 41.2 bits (95), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 21 MKLNIANPACGLQKTVEVDDEKKLLPFFERRMGAEVPGDSL 61
MKLNI PA QK +EVDDE+KL +E + EV D+L
Sbjct 1 MKLNIF-PAASCQKLIEVDDERKLRSLYEMPVATEVAADAL 40
> Hs17475469
Length=311
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 22 KLNIANPACGLQKTVEVDDEKKLLPFFER 50
K+ + PA G Q+ +EVDDE +L FE+
Sbjct 265 KMKLIFPATGCQELIEVDDEHELWTNFEK 293
> ECU03g0620
Length=414
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 38 VDDEKKLLPFFERRMGAEVPGDSLGEEFKGYLFRITGGNDKQ 79
V+ +++ F R G +VP S+G++ G + R+T D++
Sbjct 202 VESDERFSAFVSSRGGCQVPLSSIGDQAMGLVMRMTAKEDRR 243
> Hs4503409
Length=570
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 27 NPACGLQKTVEVDDEKKLLPFFERRMGAE 55
NP+C L+ + +D+E +L+ + R+ AE
Sbjct 419 NPSCMLESSNRLDEEHRLIARYAARLAAE 447
> Hs4503407
Length=743
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 27 NPACGLQKTVEVDDEKKLLPFFERRMGAE 55
NP+C L+ + +D+E +L+ + R+ AE
Sbjct 419 NPSCMLESSNRLDEEHRLIARYAARLAAE 447
> Hs14916509
Length=567
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 27 NPACGLQKTVEVDDEKKLLPFFERRMGAE 55
NP+C L+ + +D+E +L+ + R+ AE
Sbjct 416 NPSCMLESSNRLDEEHRLIARYAARLAAE 444
> CE12384
Length=853
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 80 GFPMMQGILANHRVRLCSA 98
F M+Q ++A+H+V+LCSA
Sbjct 787 AFNMLQHLVADHQVKLCSA 805
> Hs20533896
Length=5183
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 75 GNDKQGFPMMQGILANHRVRLCSAL 99
G QGF ++ ILANH ++L ++L
Sbjct 463 GKGHQGFGVLSVILANHAIKLLTSL 487
Lambda K H
0.323 0.142 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40