bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5022_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
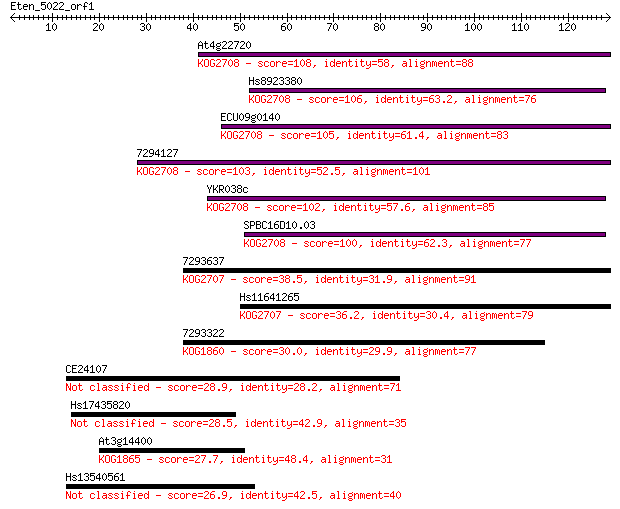
At4g22720 108 2e-24
Hs8923380 106 1e-23
ECU09g0140 105 2e-23
7294127 103 7e-23
YKR038c 102 2e-22
SPBC16D10.03 100 7e-22
7293637 38.5 0.003
Hs11641265 36.2 0.017
7293322 30.0 1.1
CE24107 28.9 2.7
Hs17435820 28.5 2.9
At3g14400 27.7 4.9
Hs13540561 26.9 8.8
> At4g22720
Length=353
Score = 108 bits (271), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 62/88 (70%), Gaps = 0/88 (0%)
Query 41 SADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQML 100
+A+ + TP +CYSLQE +F MLVEITERAMA DVL+VGGVGCN RLQ+M+
Sbjct 218 TAEEKLKNNECTPADLCYSLQETVFAMLVEITERAMAHCDKKDVLIVGGVGCNERLQEMM 277
Query 101 REMARQRGCSMGGMDERYCIDNGAMIAY 128
R M +R + D+RYCIDNGAMIAY
Sbjct 278 RTMCSERDGKLFATDDRYCIDNGAMIAY 305
> Hs8923380
Length=335
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/76 (63%), Positives = 60/76 (78%), Gaps = 0/76 (0%)
Query 52 TPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCSM 111
TPE +C+SLQE +F MLVEITERAMA S + L+VGGVGCN+RLQ+M+ M ++RG +
Sbjct 225 TPEDLCFSLQETVFAMLVEITERAMAHCGSQEALIVGGVGCNVRLQEMMATMCQERGARL 284
Query 112 GGMDERYCIDNGAMIA 127
DER+CIDNGAMIA
Sbjct 285 FATDERFCIDNGAMIA 300
> ECU09g0140
Length=331
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 51/83 (61%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 46 IPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMAR 105
I E+ +CYSLQE +F LVE+TERAMA S S +VL+VGGVGCNLRLQ+M+ MAR
Sbjct 214 IAEDEQVKRDLCYSLQETVFSALVEVTERAMAFSSSKEVLIVGGVGCNLRLQEMMGIMAR 273
Query 106 QRGCSMGGMDERYCIDNGAMIAY 128
+RG DER+CIDNG MIAY
Sbjct 274 ERGGVCYATDERFCIDNGVMIAY 296
> 7294127
Length=347
Score = 103 bits (258), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 53/101 (52%), Positives = 67/101 (66%), Gaps = 6/101 (5%)
Query 28 GGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVV 87
G + KR P E ++ + +CYSLQE +F MLVEITERAMA S++VL+V
Sbjct 218 GKRQNKRKKPQEEEVNN------YSQADLCYSLQETIFAMLVEITERAMAHCGSNEVLIV 271
Query 88 GGVGCNLRLQQMLREMARQRGCSMGGMDERYCIDNGAMIAY 128
GGVGCN RLQ+M+R M +RG + DERYCIDNG MIA+
Sbjct 272 GGVGCNERLQEMMRIMCEERGGKLFATDERYCIDNGLMIAH 312
> YKR038c
Length=421
Score = 102 bits (253), Expect = 2e-22, Method: Composition-based stats.
Identities = 49/86 (56%), Positives = 63/86 (73%), Gaps = 1/86 (1%)
Query 43 DSSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLRE 102
D + E+ +T E +CYSLQE LF MLVEITERAMA +S+ VL+VGGVGCN+RLQ+M+ +
Sbjct 300 DKTTGEQKVTVEDLCYSLQENLFAMLVEITERAMAHVNSNQVLIVGGVGCNVRLQEMMAQ 359
Query 103 MARQRG-CSMGGMDERYCIDNGAMIA 127
M + R + D R+CIDNG MIA
Sbjct 360 MCKDRANGQVHATDNRFCIDNGVMIA 385
> SPBC16D10.03
Length=346
Score = 100 bits (249), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 48/77 (62%), Positives = 57/77 (74%), Gaps = 0/77 (0%)
Query 51 LTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLRLQQMLREMARQRGCS 110
+T + +CYSLQE F MLVEITERAMA + VL+VGGVGCN RLQQM+ EM+ RG
Sbjct 235 VTKQDLCYSLQETGFAMLVEITERAMAHIRADSVLIVGGVGCNERLQQMMAEMSSDRGAD 294
Query 111 MGGMDERYCIDNGAMIA 127
+ DER+CIDNG MIA
Sbjct 295 VFSTDERFCIDNGIMIA 311
> 7293637
Length=409
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 48/102 (47%), Gaps = 11/102 (10%)
Query 38 SRESADSSIPEELLTPE-SICYSLQEYLFGMLVEITERAMA---LSH-------SSDVLV 86
+RE A+ + P+ +++ C L + L+ T+RA+ L H +++
Sbjct 257 ARERAERTPPDGVISNYGDFCAGLLRSVSRHLMHRTQRAIEYCLLPHRQLFGDTPPTLVM 316
Query 87 VGGVGCNLRLQQMLREMARQRGCSMGGMDERYCIDNGAMIAY 128
GGV N + + +A Q GC +RYC DNG MIA+
Sbjct 317 SGGVANNDAIYANIEHLAAQYGCRSFRPSKRYCSDNGVMIAW 358
> Hs11641265
Length=439
Score = 36.2 bits (82), Expect = 0.017, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 8/87 (9%)
Query 50 LLTPESICYSLQEYLFGMLVEITERAMALSHSSDVL--------VVGGVGCNLRLQQMLR 101
L + I ++Q + LV+ T RA+ D+L GGV N +++ L
Sbjct 304 LSSAADIAATVQHTMACHLVKRTHRAILFCKQRDLLPQNNAVLVASGGVASNFYIRRALE 363
Query 102 EMARQRGCSMGGMDERYCIDNGAMIAY 128
+ C++ R C DNG MIA+
Sbjct 364 ILTNATQCTLLCPPPRLCTDNGIMIAW 390
> 7293322
Length=2122
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 38/79 (48%), Gaps = 7/79 (8%)
Query 38 SRESAD--SSIPEELLTPESICYSLQEYLFGMLVEITERAMALSHSSDVLVVGGVGCNLR 95
SR SAD + +P EL E+ + YL +++I+ER SH D R
Sbjct 243 SRSSADQETPLPHELRN-ETALHMTMSYLMHEIMDISERQDPQSHMGDWFHFVWD----R 297
Query 96 LQQMLREMARQRGCSMGGM 114
+ + +E+ +Q CS+G +
Sbjct 298 TRSIRKEITQQELCSLGAV 316
> CE24107
Length=1257
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 32/71 (45%), Gaps = 0/71 (0%)
Query 13 EVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLTPESICYSLQEYLFGMLVEIT 72
E A + + +R + G R + SRE D + P EL E + + + LFG V +
Sbjct 475 EHARQDFEHVRANQLGDNFYIRSHFSREKRDKASPLELSINEGDIFHVTDTLFGGTVGLW 534
Query 73 ERAMALSHSSD 83
+ A S S +
Sbjct 535 QAARVYSSSEN 545
> Hs17435820
Length=145
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 14 VAEEMAKRMRTKRSGGQLRKRGNPSRESAD--SSIPE 48
V + + R +R G LR + NPSRE+ D SS+P
Sbjct 51 VTDSHSTRKTHRRIGQVLRVQANPSREAVDCSSSVPN 87
> At3g14400
Length=661
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 4/35 (11%)
Query 20 KRMRTKRSGGQ----LRKRGNPSRESADSSIPEEL 50
KR+RT++SGG+ L++ + S S IPEEL
Sbjct 593 KRLRTEQSGGEDGSDLKRLIEDVKSSLKSQIPEEL 627
> Hs13540561
Length=151
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 13 EVAEEMAKRMRTKRSGGQLRKRGNPSRESADSSIPEELLT 52
E EEM +R R R+G QL+++ N E S+ EEL T
Sbjct 81 EQLEEMEERQRQLRNGVQLQQQKNKEMEQLRLSLAEELST 120
Lambda K H
0.319 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40