bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_5018_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
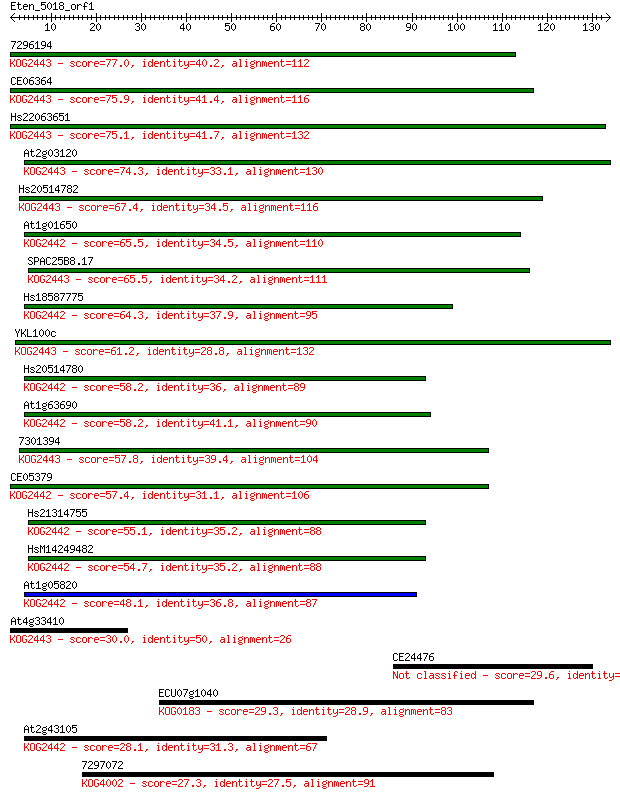
7296194 77.0 9e-15
CE06364 75.9 2e-14
Hs22063651 75.1 3e-14
At2g03120 74.3 5e-14
Hs20514782 67.4 7e-12
At1g01650 65.5 2e-11
SPAC25B8.17 65.5 3e-11
Hs18587775 64.3 6e-11
YKL100c 61.2 6e-10
Hs20514780 58.2 4e-09
At1g63690 58.2 5e-09
7301394 57.8 5e-09
CE05379 57.4 6e-09
Hs21314755 55.1 4e-08
HsM14249482 54.7 5e-08
At1g05820 48.1 4e-06
At4g33410 30.0 1.4
CE24476 29.6 1.7
ECU07g1040 29.3 1.9
At2g43105 28.1 5.1
7297072 27.3 7.2
> 7296194
Length=389
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 17/112 (15%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGD+VIPG+ IA+ LRFD D +R + YF L
Sbjct 264 ASNFAMLGLGDIVIPGIFIALLLRFD-----------------DSKKRKTRIYFYSTLIA 306
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEE 112
Y LGLL T VM +H QPALLYLVP C+ + A + G++K + AY++
Sbjct 307 YFLGLLATIFVMHVFKHAQPALLYLVPACMGTPLLVALIRGELKVLFAYEDH 358
> CE06364
Length=468
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 67/120 (55%), Gaps = 9/120 (7%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A ++S+LGLGD+VIPG+ IA+ RFD + Q A +KA + R+ YF V +
Sbjct 343 ASKHSMLGLGDIVIPGIFIALLRRFDYRVVQ--TTAESKAPQGSLKGRY---YFVVTVVA 397
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKE----EDEEG 116
Y GL T VM + + QPALLYLVP CLF A + G++ + Y E ++EE
Sbjct 398 YMAGLFITMAVMHHFKAAQPALLYLVPCCLFVPLLLAVIRGELSALWNYDESRHVDNEEN 457
> Hs22063651
Length=377
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 73/142 (51%), Gaps = 30/142 (21%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
A +++LGLGDVVIPG+ IA+ LRFD+ L ++N T YF A
Sbjct 255 ANNFAMLGLGDVVIPGIFIALLLRFDISL---KKNTHT--------------YFYTSFAA 297
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKE--------- 111
Y GL T +M +H QPALLYLVP C+ A G+V E+ +Y+E
Sbjct 298 YIFGLGLTIFIMHIFKHAQPALLYLVPACIGFPVLVALAKGEVTEMFSYEESNPKDPAAV 357
Query 112 -EDEEGSSILRASALETTEKKE 132
E +EG+ ASA + EKKE
Sbjct 358 TESKEGT---EASASKGLEKKE 376
> At2g03120
Length=344
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 65/130 (50%), Gaps = 18/130 (13%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YS+LGLGD+VIPG+ +A+ LRFD+ ++ Q YF Y +
Sbjct 232 YSMLGLGDIVIPGIFVALALRFDVSRRRQPQ------------------YFTSAFIGYAV 273
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSILRAS 123
G++ T +VM + Q QPALLY+VP + L NG +K +LA+ E E ++ +
Sbjct 274 GVILTIVVMNWFQAAQPALLYIVPAVIGFLASHCIWNGDIKPLLAFDESKTEEATTDESK 333
Query 124 ALETTEKKEN 133
E K +
Sbjct 334 TSEEVNKAHD 343
> Hs20514782
Length=385
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 61/117 (52%), Gaps = 1/117 (0%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKF-YFCVVLAFY 61
+S+LG+GD+V+PG+L+ LR+D + Q ++ +I R K YF L Y
Sbjct 264 HFSMLGIGDIVMPGLLLCFVLRYDNYKKQASGDSCGAPGPANISGRMQKVSYFHCTLIGY 323
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSS 118
+GLLT + + QPALLYLVP+ L L A L G ++ + + + SS
Sbjct 324 FVGLLTATVASRIHRAAQPALLYLVPFTLLPLLTMAYLKGDLRRMWSEPFHSKSSSS 380
> At1g01650
Length=491
Score = 65.5 bits (158), Expect = 2e-11, Method: Composition-based stats.
Identities = 38/111 (34%), Positives = 56/111 (50%), Gaps = 17/111 (15%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YSI+G GD+++PG+L+ LR+D ++R YF ++ Y L
Sbjct 384 YSIIGFGDIILPGLLVTFALRYDWL----------------ANKRLKSGYFLGTMSAYGL 427
Query 64 GLLTTGLVM-LYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEED 113
GLL T + + L H QPALLY+VP+ L +LF G +K + E D
Sbjct 428 GLLITYIALNLMDGHGQPALLYIVPFILGTLFVLGHKRGDLKTLWTTGEPD 478
> SPAC25B8.17
Length=295
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 57/111 (51%), Gaps = 10/111 (9%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
S+LGLGD+V+PG+++A+ RFDL Y + K H YF Y LG
Sbjct 184 SMLGLGDIVMPGLMLALMYRFDLHYYINSTSQPKK------HST----YFRNTFIAYGLG 233
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEE 115
L T + Y + QPALLYL P C+ + A ++K + +++ E E+
Sbjct 234 LGVTNFALYYFKAAQPALLYLSPACIVAPLLTAWYRDELKTLFSFRSETED 284
> Hs18587775
Length=684
Score = 64.3 bits (155), Expect = 6e-11, Method: Composition-based stats.
Identities = 36/95 (37%), Positives = 47/95 (49%), Gaps = 18/95 (18%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+SILG GD+V+PG L+A C RFD+ + +Q YF Y +
Sbjct 441 FSILGFGDIVVPGFLVAYCCRFDVQVCSRQ------------------IYFVACTVAYAV 482
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAA 98
GLL T + M+ Q QPALLYLV L + AA
Sbjct 483 GLLVTFMAMVLMQMGQPALLYLVSSTLLTSLAVAA 517
> YKL100c
Length=587
Score = 61.2 bits (147), Expect = 6e-10, Method: Composition-based stats.
Identities = 38/132 (28%), Positives = 63/132 (47%), Gaps = 7/132 (5%)
Query 2 LQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFY 61
+SILGLGD+ +PG+ IAMC ++D++ + + ++ + YF + Y
Sbjct 402 FNFSILGLGDIALPGMFIAMCYKYDIWKWHLDHDDTEFHF---LNWSYVGKYFITAMVSY 458
Query 62 ELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEVLAYKEEDEEGSSILR 121
L++ + + QPALLY+VP L S A N K+ ++ + E L+
Sbjct 459 VASLVSAMVSLSIFNTAQPALLYIVPSLLISTILVACWNKDFKQFWNFQYDTIEVDKSLK 518
Query 122 ASALETTEKKEN 133
+ EKKEN
Sbjct 519 ----KAIEKKEN 526
> Hs20514780
Length=564
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 32/89 (35%), Positives = 45/89 (50%), Gaps = 18/89 (20%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
+S+LG GD+++PG+L+A C RF DI + + YF Y +
Sbjct 386 FSLLGFGDILVPGLLVAYCHRF------------------DIQVQSSRVYFVACTIAYGV 427
Query 64 GLLTTGLVMLYAQHPQPALLYLVPYCLFS 92
GLL T + + Q QPALLYLVP L +
Sbjct 428 GLLVTFVALALMQRGQPALLYLVPCTLVT 456
> At1g63690
Length=519
Score = 58.2 bits (139), Expect = 5e-09, Method: Composition-based stats.
Identities = 37/91 (40%), Positives = 48/91 (52%), Gaps = 17/91 (18%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
YSI+G GD+++PG+LIA LR+D A K L YF + Y L
Sbjct 431 YSIIGFGDILLPGLLIAFALRYDWL--------ANKTLRTG--------YFIWAMVAYGL 474
Query 64 GLLTTGLVM-LYAQHPQPALLYLVPYCLFSL 93
GLL T + + L H QPALLY+VP+ L L
Sbjct 475 GLLITYVALNLMDGHGQPALLYIVPFTLGKL 505
> 7301394
Length=417
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 41/107 (38%), Positives = 60/107 (56%), Gaps = 7/107 (6%)
Query 3 QYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAA---TKALAVDIHQRFPKFYFCVVLA 59
+S+LGLGDVV+PG+L+ LR+D Y+K Q T + + R F+ C +L
Sbjct 299 HFSMLGLGDVVMPGLLLCFVLRYDA--YKKAQGVTSDPTLSPPRGVGSRLTYFH-CSLLG 355
Query 60 FYELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEV 106
++ LGLLT + + QPALLYLVP+ L L A L G ++ +
Sbjct 356 YF-LGLLTATVSSEVFKAAQPALLYLVPFTLLPLLLMAYLKGDLRRM 401
> CE05379
Length=652
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 51/106 (48%), Gaps = 17/106 (16%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAF 60
Q++ILGLGD+V+PG L+A C + F +R Y + +
Sbjct 506 GFQFTILGLGDIVMPGYLVAHCFTMNGF-----------------SERVRLIYGFISVVG 548
Query 61 YELGLLTTGLVMLYAQHPQPALLYLVPYCLFSLFGAAALNGKVKEV 106
Y +GL+ T L + + QPAL+YLVP LF + A G+ ++
Sbjct 549 YGIGLIVTFLALALMKTAQPALIYLVPSTLFPIIMLALCRGEFLKI 594
> Hs21314755
Length=520
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 18/88 (20%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
SILG GD+++PG+LIA C RFD+ Q ++ Y+ Y +G
Sbjct 406 SILGFGDIIVPGLLIAYCRRFDV------QTGSS------------YIYYVSSTVAYAIG 447
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFS 92
++ T +V++ + QPALLYLVP L +
Sbjct 448 MILTFVVLVLMKKGQPALLYLVPCTLIT 475
> HsM14249482
Length=409
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 18/88 (20%)
Query 5 SILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYELG 64
SILG GD+++PG+LIA C RFD+ Q ++ Y+ Y +G
Sbjct 295 SILGFGDIIVPGLLIAYCRRFDV------QTGSS------------YIYYVSSTVAYAIG 336
Query 65 LLTTGLVMLYAQHPQPALLYLVPYCLFS 92
++ T +V++ + QPALLYLVP L +
Sbjct 337 MILTFVVLVLMKKGQPALLYLVPCTLIT 364
> At1g05820
Length=441
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 32/88 (36%), Positives = 43/88 (48%), Gaps = 17/88 (19%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKFYFCVVLAFYEL 63
Y+++G GD++ PG+LI RFD K+ N YF ++ Y L
Sbjct 335 YNMIGFGDILFPGLLICFIFRFD-----KENNKGVS-----------NGYFPWLMFGYGL 378
Query 64 GLLTTGL-VMLYAQHPQPALLYLVPYCL 90
GL T L + + H QPALLYLVP L
Sbjct 379 GLFLTYLGLYVMNGHGQPALLYLVPCTL 406
> At4g33410
Length=311
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 1 ALQYSILGLGDVVIPGVLIAMCLRFD 26
A + +LGLGD+ IP +L+A+ L FD
Sbjct 222 ASDFMMLGLGDMAIPAMLLALVLCFD 247
> CE24476
Length=553
Score = 29.6 bits (65), Expect = 1.7, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query 86 VPYCLF---SLFGAAALNGKVKEVLAYKEEDEEGSSILRASALETTE 129
+P LF F +A L+ E A KEEDE G S LR S+L T +
Sbjct 415 LPTALFGHLKAFQSALLSTVKYEPPARKEEDEAGFSTLRLSSLATED 461
> ECU07g1040
Length=224
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 39/86 (45%), Gaps = 7/86 (8%)
Query 34 QNAATKALAVDIHQRFPKFY---FCVVLAFYELGLLTTGLVMLYAQHPQPALLYLVPYCL 90
+N+ + +A+D R Y + VV G+ T +L+ P+P L P
Sbjct 95 KNSTGEDIAIDQLARMLSEYKQRYTVVYHQRPFGVRT----VLFGLEPRPVAYVLEPDGN 150
Query 91 FSLFGAAALNGKVKEVLAYKEEDEEG 116
FS F A+ K ++V Y E+ E+G
Sbjct 151 FSEFFCGAIGQKSQKVCEYLEKGEKG 176
> At2g43105
Length=543
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 30/75 (40%), Gaps = 8/75 (10%)
Query 4 YSILGLGDVVIPGVLIAMCLRFDLFLYQKQQNAATKALAVDIHQRFPKF--------YFC 55
Y ++G GD++ PG+LI+ R LA R+ K YF
Sbjct 433 YDMIGFGDILFPGLLISFASRVSFSTILSSNPLPRLILAPLFDLRYDKIKKRVISNGYFL 492
Query 56 VVLAFYELGLLTTGL 70
+ Y +GLL T L
Sbjct 493 WLTIGYGIGLLLTYL 507
> 7297072
Length=276
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 52/101 (51%), Gaps = 11/101 (10%)
Query 17 VLIAMCLRFDLFLYQKQQNAATKAL-------AVDIHQRFPKFYFCVVLAFYELGLLTTG 69
++I + + L ++ QQ+A +K + A+ +HQR P F F L +
Sbjct 60 LVIFFTVSYVLPIFTSQQSAFSKVMLANAAISALRLHQRLPAFAFSREF-LARLFAEDSC 118
Query 70 LVMLYAQ---HPQPALLYLVPYCLFSLFGAAALNGKVKEVL 107
M+Y+ + +P+LL L+P L+S+ A++ + K+ +++
Sbjct 119 HYMMYSLIFFNIRPSLLVLIPVLLYSVLHASSYSLKLLDLI 159
Lambda K H
0.325 0.140 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40