bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4999_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
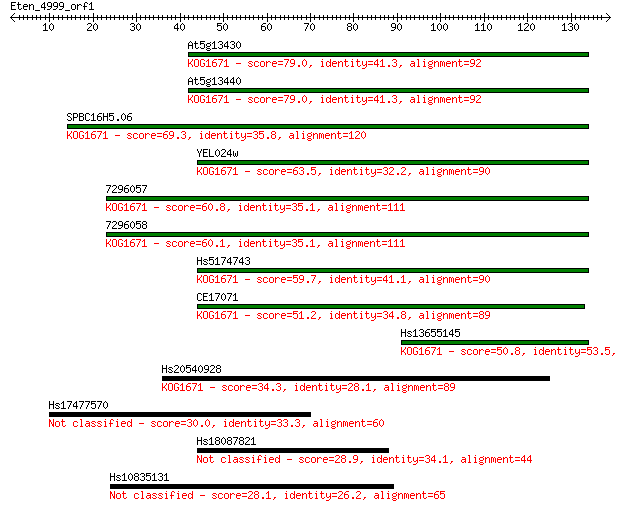
At5g13430 79.0 3e-15
At5g13440 79.0 3e-15
SPBC16H5.06 69.3 2e-12
YEL024w 63.5 1e-10
7296057 60.8 8e-10
7296058 60.1 1e-09
Hs5174743 59.7 2e-09
CE17071 51.2 6e-07
Hs13655145 50.8 8e-07
Hs20540928 34.3 0.075
Hs17477570 30.0 1.3
Hs18087821 28.9 2.8
Hs10835131 28.1 5.1
> At5g13430
Length=274
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 42 AHPDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTE 101
+ D R G P +R F YF+ + F+ AS++R L+ K++ SKD++A + E
Sbjct 94 VYDDHNHERYPPGDPSKRAFAYFVLSGGRFVYASVLRLLVLKLIVSMSASKDVLALASLE 153
Query 102 IDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+DL IE GTT KWRGKPVF+R RT ++I+
Sbjct 154 VDLGSIEPGTTVTVKWRGKPVFIRRRTEDDIK 185
> At5g13440
Length=274
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 42 AHPDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTE 101
+ D R G P +R F YF+ + F+ AS++R L+ K++ SKD++A + E
Sbjct 94 VYDDHNHERYPPGDPSKRAFAYFVLSGGRFVYASVLRLLVLKLIVSMSASKDVLALASLE 153
Query 102 IDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+DL IE GTT KWRGKPVF+R RT ++I+
Sbjct 154 VDLGSIEPGTTVTVKWRGKPVFIRRRTEDDIK 185
> SPBC16H5.06
Length=228
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/127 (33%), Positives = 63/127 (49%), Gaps = 7/127 (5%)
Query 14 SVGRFAPDNFRAA---GYAENAPNPTSI----NSDAHPDFRDYRLGSGSPDRRPFMYFLS 66
S+ R P + A+ G P TSI +S PDF +Y+ S R Y +
Sbjct 14 SLRRLLPVSSTASSLKGSMMTIPKFTSIRTYTDSPEMPDFSEYQTKSTGDRSRVISYAMV 73
Query 67 ASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRH 126
+ + A+ ++ + + W S D++A E+DL I G V KW+GKPVF+RH
Sbjct 74 GTMGALTAAGAQATVHDFLASWSASADVLAMSKAEVDLSKIPEGKNLVVKWQGKPVFIRH 133
Query 127 RTPEEIE 133
RTPEEI+
Sbjct 134 RTPEEIQ 140
> YEL024w
Length=215
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 50/91 (54%), Gaps = 1/91 (1%)
Query 44 PDFRDYRLGSGSPDR-RPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEI 102
P+F D + D+ R + YF+ + + ++ +S + + + D++A E+
Sbjct 37 PNFDDVLKENNDADKGRSYAYFMVGAMGLLSSAGAKSTVETFISSMTATADVLAMAKVEV 96
Query 103 DLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+L I +G V KW+GKPVF+RHRTP EI+
Sbjct 97 NLAAIPLGKNVVVKWQGKPVFIRHRTPHEIQ 127
> 7296057
Length=260
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 58/121 (47%), Gaps = 10/121 (8%)
Query 23 FRAAGYAENAPNPTSINSDAH-PDFRDYRLGS---------GSPDRRPFMYFLSASYFFI 72
+A+G A N+ ++D PDF YR S + +R+ F Y + +
Sbjct 21 LKASGVAVNSMANRQAHTDLQVPDFSAYRRESVKDSRRRNDTAEERKAFSYLMVGAGAVG 80
Query 73 GASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEI 132
GA + L+ + S +++A EI L I G + FKWRGKP+F+RHRT EI
Sbjct 81 GAYAAKGLVNTFIGSMSASAEVLAMAKIEIKLSDIPEGKSVTFKWRGKPLFIRHRTAAEI 140
Query 133 E 133
E
Sbjct 141 E 141
> 7296058
Length=230
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 58/121 (47%), Gaps = 10/121 (8%)
Query 23 FRAAGYAENAPNPTSINSDAH-PDFRDYRLGS---------GSPDRRPFMYFLSASYFFI 72
+A+G A N+ ++D PDF YR S + +R+ F Y + +
Sbjct 21 LKASGVAVNSMANRQAHTDLQVPDFSAYRRESVKDSRRRNDTAEERKAFSYLMVGAGAVG 80
Query 73 GASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEI 132
GA + L+ + S +++A EI L I G + FKWRGKP+F+RHRT EI
Sbjct 81 GAYAAKGLVNTFIGSMSASAEVLAMAKIEIKLSDIPEGKSVTFKWRGKPLFIRHRTAAEI 140
Query 133 E 133
E
Sbjct 141 E 141
> Hs5174743
Length=274
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 49/100 (49%), Gaps = 10/100 (10%)
Query 44 PDFRDYR----LGS------GSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF +YR L S S R+ F Y ++ A ++ + + V S D
Sbjct 86 PDFSEYRRLEVLDSTKSSRESSEARKGFSYLVTGVTTVGVAYAAKNAVTQFVSSMSASAD 145
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
++A EI L I G FKWRGKP+FVRHRT +EIE
Sbjct 146 VLALAKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTQKEIE 185
> CE17071
Length=276
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 11/100 (11%)
Query 44 PDFRDYRLGSGSPDRRPFM-------YFLSASYFFIGASMM----RSLLCKMVHFWWVSK 92
PD +YR S + P +A Y+ G + + ++ +V + ++
Sbjct 86 PDMSNYRRDSTKNTQVPARDTEDQRRALPTALYYGAGGVLSLWAGKEVVQTLVSYKAMAA 145
Query 93 DLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEI 132
D A + EI++ I G T F+WRGKPVFV+HRT EI
Sbjct 146 DQRALASIEINMADIPEGKTKTFEWRGKPVFVKHRTKAEI 185
> Hs13655145
Length=135
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 91 SKDLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
S D++A EI L I G FKWRGKP+FVRHRT +EI+
Sbjct 4 SADVLALAKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTQKEIK 46
> Hs20540928
Length=160
Score = 34.3 bits (77), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 39/96 (40%), Gaps = 7/96 (7%)
Query 36 TSINSDAHPDFRDYRLGSGSPD-------RRPFMYFLSASYFFIGASMMRSLLCKMVHFW 88
T IN PD+ + G+ R+ F Y ++A+ ++++ + V
Sbjct 22 TDINVPDFPDYCCIEIFDGTKSSKEDGEARKGFSYLVTATTAVGVTYAAKNVISQFVSSI 81
Query 89 WVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFV 124
S D A I L I G FKWRGK +F+
Sbjct 82 SASADESAVSKIAIKLFDIPEGKNMAFKWRGKSLFI 117
> Hs17477570
Length=1361
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 10 PAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRL--------GSGSPDRRPF 61
P SVG ++P GY+E+ P N+ AH ++R + + + D+RPF
Sbjct 703 PTGPSVGMYSP------GYSEHIPLGKFYNNRAHSNYRAGMIIDNGVKTTEASAKDKRPF 756
Query 62 MYFLSASY 69
+ +SA Y
Sbjct 757 LSIISARY 764
> Hs18087821
Length=116
Score = 28.9 bits (63), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 44 PDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHF 87
PD+RD GSG D+ + + A++ F+G S + LL K++
Sbjct 49 PDYRDPPDGSGRLDQLLSLSMVWANHLFLGCSYNKDLLDKVMEM 92
> Hs10835131
Length=420
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 28/65 (43%), Gaps = 13/65 (20%)
Query 24 RAAGYAENAPNPTSINSDAHPDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCK 83
R AG P + +D H R++ F+ + A+ FI ++ SL+CK
Sbjct 317 REAGLWSEWSQPIYVGNDEHKPLREW-----------FVIVIMATICFI--LLILSLICK 363
Query 84 MVHFW 88
+ H W
Sbjct 364 ICHLW 368
Lambda K H
0.322 0.137 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40