bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4998_orf3
Length=82
Score E
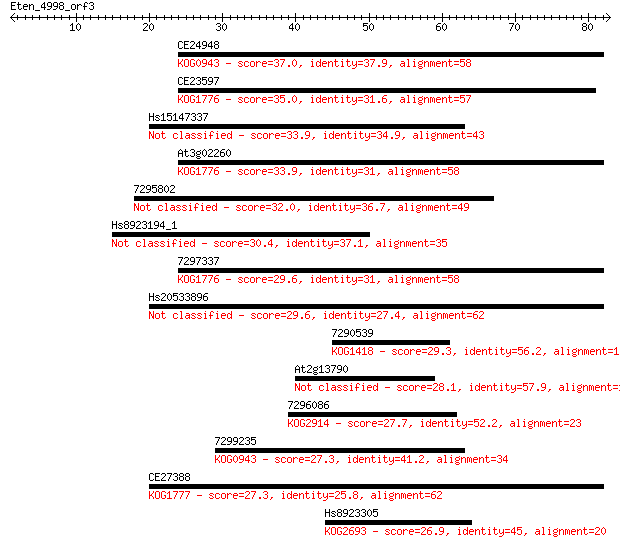
Sequences producing significant alignments: (Bits) Value
CE24948 37.0 0.008
CE23597 35.0 0.037
Hs15147337 33.9 0.082
At3g02260 33.9 0.082
7295802 32.0 0.27
Hs8923194_1 30.4 0.92
7297337 29.6 1.3
Hs20533896 29.6 1.5
7290539 29.3 1.7
At2g13790 28.1 4.4
7296086 27.7 5.7
7299235 27.3 6.7
CE27388 27.3 6.8
Hs8923305 26.9 9.1
> CE24948
Length=2944
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 8/66 (12%)
Query 24 CTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCK----RPQRVSVTS-CGSF--- 75
C+ TG Q + CKTC + CCCS C C + R +R S T+ C +
Sbjct 1204 CSFTWTGEDHINQDIYECKTCGLTGSLCCCSECALTCHRNHDCRLKRTSPTAYCDCWEKS 1263
Query 76 AANCAA 81
+ NC A
Sbjct 1264 SCNCKA 1269
> CE23597
Length=1131
Score = 35.0 bits (79), Expect = 0.037, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 24/57 (42%), Gaps = 1/57 (1%)
Query 24 CTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCKRPQRVSVTSCGSFAANCA 80
CT TG A Q + C TC+ C S C C R ++ + GSF +C
Sbjct 794 CTYKSTGRAYVTQHWYNCYTCNMMESEGVC-SVCAINCHRGHDLAYSKKGSFFCDCG 849
> Hs15147337
Length=2799
Score = 33.9 bits (76), Expect = 0.082, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 18/43 (41%), Gaps = 0/43 (0%)
Query 20 CAFACTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCK 62
C C+ TG Q F C+TC CCC+ C C K
Sbjct 1175 CNDTCSFTWTGAEHINQDIFECRTCGLLESLCCCTECARVCHK 1217
> At3g02260
Length=5079
Score = 33.9 bits (76), Expect = 0.082, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 1/58 (1%)
Query 24 CTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCKRPQRVSVTSCGSFAANCAA 81
CT +G+ Q + C TC + CCS C C R RV + F +C A
Sbjct 1575 CTFTSSGSNFMEQHWYFCYTCDLTVSKGCCSV-CAKVCHRGHRVVYSRSSRFFCDCGA 1631
> 7295802
Length=773
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 22/54 (40%), Gaps = 15/54 (27%)
Query 18 PNCAFACTSGGTGNALCMQRRFGCKTCSSSSCCC-----CCSSCCCCCCKRPQR 66
P C C G+ N L TC +S C C C+ C C CCK P +
Sbjct 523 PKCR--CGISGSSNTL--------TTCRNSRCPCYKSYNSCAGCHCVCCKNPHK 566
> Hs8923194_1
Length=519
Score = 30.4 bits (67), Expect = 0.92, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 15 DFLPNCAFACTSGGTGNALCMQRRFGCKTCSSSSC 49
D +CA +C GG + L +R C CSS C
Sbjct 302 DGAKDCACSCHEGGPDSKLKKSKRRSCSHCSSKVC 336
> 7297337
Length=5322
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 23/58 (39%), Gaps = 1/58 (1%)
Query 24 CTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCKRPQRVSVTSCGSFAANCAA 81
CT T Q + C TC+ + CS C C K VS G+F +C A
Sbjct 1796 CTFSQTQKEFMNQHWYHCHTCNMINTVGVCSVCARVCHK-GHDVSYAKYGNFFCDCGA 1852
> Hs20533896
Length=5183
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 22/62 (35%), Gaps = 1/62 (1%)
Query 20 CAFACTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCKRPQRVSVTSCGSFAANC 79
C CT T Q + C TC C + C C + +S GSF +C
Sbjct 1658 CNKLCTFTITQKEFMNQHWYHCHTCKMVDGVGVC-TVCAKVCHKDHEISYAKYGSFFCDC 1716
Query 80 AA 81
A
Sbjct 1717 GA 1718
> 7290539
Length=592
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 9/16 (56%), Positives = 11/16 (68%), Gaps = 0/16 (0%)
Query 45 SSSSCCCCCSSCCCCC 60
S + CCC CS+C CC
Sbjct 246 SKALCCCLCSNCGYCC 261
> At2g13790
Length=520
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 13/21 (61%), Gaps = 2/21 (9%)
Query 40 GCKTCSSS--SCCCCCSSCCC 58
G CS+S SCC C +S CC
Sbjct 142 GANDCSNSRGSCCRCSTSICC 162
> 7296086
Length=305
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 13/23 (56%), Gaps = 0/23 (0%)
Query 39 FGCKTCSSSSCCCCCSSCCCCCC 61
FGC+TC S C CS CC C
Sbjct 4 FGCRTCKSQPDCEGCSPKCCSPC 26
> 7299235
Length=2165
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 16/34 (47%), Gaps = 0/34 (0%)
Query 29 TGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCK 62
TG Q F CKTC + CCC+ C C K
Sbjct 1221 TGADHINQNIFECKTCGLTGSLCCCTECARVCHK 1254
> CE27388
Length=936
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 26/62 (41%), Gaps = 4/62 (6%)
Query 20 CAFACTSGGTGNALCMQRRFGCKTCSSSSCCCCCSSCCCCCCKRPQRVSVTSCGSFAANC 79
C F +S N+ M + C TC+++ C++ C C R V + F +C
Sbjct 845 CLFKVSSN---NSFPMHNFYRCTTCNTTERNAICTN-CIRTCHRGHSVELVRFDRFFCDC 900
Query 80 AA 81
A
Sbjct 901 GA 902
> Hs8923305
Length=626
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 9/20 (45%), Positives = 12/20 (60%), Gaps = 0/20 (0%)
Query 44 CSSSSCCCCCSSCCCCCCKR 63
C++ +C SCCCC C R
Sbjct 598 CTTWACWAAGPSCCCCPCTR 617
Lambda K H
0.331 0.133 0.503
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1159278568
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40