bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4995_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
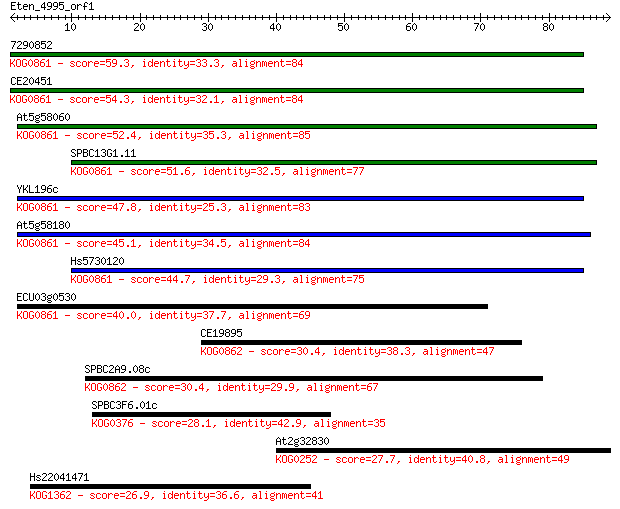
7290852 59.3 1e-09
CE20451 54.3 5e-08
At5g58060 52.4 2e-07
SPBC13G1.11 51.6 3e-07
YKL196c 47.8 5e-06
At5g58180 45.1 3e-05
Hs5730120 44.7 4e-05
ECU03g0530 40.0 0.001
CE19895 30.4 0.75
SPBC2A9.08c 30.4 0.92
SPBC3F6.01c 28.1 3.7
At2g32830 27.7 5.5
Hs22041471 26.9 9.1
> 7290852
Length=199
Score = 59.3 bits (142), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 44/84 (52%), Gaps = 0/84 (0%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYP 60
+FSF+ R TV E + F KT + R +Q V+ + V+ ++LA + + D EYP
Sbjct 28 SFSFFQRGTVNEFMTFASKTIVERTQPALRQSVKQDAYMCHVYVRADNLAGVLIADHEYP 87
Query 61 RRVAFACLNEVYLQFTQSVPASKW 84
RVA + ++ FT V A +W
Sbjct 88 HRVAHTLITKILDDFTAKVSADQW 111
> CE20451
Length=201
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 0/84 (0%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYP 60
+FSF+ R +V+E + F K + R + ++ V+ + + L+A+ V D EY
Sbjct 29 SFSFFQRGSVQEFMTFTAKLLVERSGLGARSSVKENEYLVHCYVRNDGLSAVCVTDAEYQ 88
Query 61 RRVAFACLNEVYLQFTQSVPASKW 84
+RVA + L V FT VPA++W
Sbjct 89 QRVAMSFLGRVLDDFTTRVPATQW 112
> At5g58060
Length=199
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 47/85 (55%), Gaps = 2/85 (2%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPR 61
F ++ R++VKE + FV +T R P + +Q V+HE + N L A+ D YP
Sbjct 30 FGYFQRSSVKEFVVFVGRTVASRTPPSQRQSVQHEEYKVHAYNR-NGLCAVGFMDDHYPV 88
Query 62 RVAFACLNEVYLQFTQSVPASKWKS 86
R AF+ LN+V ++ +S S W+S
Sbjct 89 RSAFSLLNQVLDEYQKSFGES-WRS 112
> SPBC13G1.11
Length=197
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 38/77 (49%), Gaps = 0/77 (0%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACLN 69
+ E + F KT R +Q+VE V+ + L + DKEYP RVA+ LN
Sbjct 36 IGEFMNFFTKTVAERTNPGQRQDVEQSNYVFHVYNRSDGLCGVIASDKEYPLRVAYTLLN 95
Query 70 EVYLQFTQSVPASKWKS 86
++ +F P +KW+S
Sbjct 96 KILDEFLTKNPRTKWES 112
> YKL196c
Length=200
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPR 61
F F+ R++V + + F +T R +Q +E V+ + + + DKEYP
Sbjct 28 FGFFERSSVGQFMTFFAETVASRTGAGQRQSIEEGNYIGHVYARSEGICGVLITDKEYPV 87
Query 62 RVAFACLNEVYLQFTQSVPASKW 84
R A+ LN++ ++ + P +W
Sbjct 88 RPAYTLLNKILDEYLVAHPKEEW 110
> At5g58180
Length=199
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPR 61
FSFY R+ +E + F+ +T R P +Q V+HE + N L A+ D YP
Sbjct 33 FSFY-RSNFEEFIVFIARTVARRTPPGQRQSVKHEEYKVHAYNI-NGLCAVGFMDDHYPV 90
Query 62 RVAFACLNEVYLQFTQSVPASKWK 85
R AF+ LN+V L Q W+
Sbjct 91 RSAFSLLNQV-LDVYQKDYGDTWR 113
> Hs5730120
Length=198
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACLN 69
V+E + F + + R ++ V+ + V+ +SLA + + D EYP RVAF L
Sbjct 36 VQEFMTFTSQLIVERSSKGTRASVKEQDYLCHVYVRNDSLAGVVIADNEYPSRVAFTLLE 95
Query 70 EVYLQFTQSVPASKW 84
+V +F++ V W
Sbjct 96 KVLDEFSKQVDRIDW 110
> ECU03g0530
Length=176
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 33/73 (45%), Gaps = 4/73 (5%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRY----PNSLAALAVGDK 57
FSF+ R V+E L F+ K +L QE HE +R N LA +A D
Sbjct 26 FSFFTRGKVRETLMFISKELAEKLETYEFQEYTHEFNDKKTYRLFSLVHNDLAYIACADD 85
Query 58 EYPRRVAFACLNE 70
+YP VA L E
Sbjct 86 DYPGIVALKLLQE 98
> CE19895
Length=214
Score = 30.4 bits (67), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 5/49 (10%)
Query 29 SQQEVEHEGLCAFVFRY--PNSLAALAVGDKEYPRRVAFACLNEVYLQF 75
+QQ VE FVF Y ++ AL + D+ +PR+VAF L+++ +F
Sbjct 50 AQQSVES---GPFVFHYIIVQNICALVLCDRNFPRKVAFQYLSDIGQEF 95
> SPBC2A9.08c
Length=209
Score = 30.4 bits (67), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 1/68 (1%)
Query 12 EGLKFVVKTAIPRLPVNSQQEVEHE-GLCAFVFRYPNSLAALAVGDKEYPRRVAFACLNE 70
E K K + RL S++ E G F + N + L + ++ YPR++AF+ L E
Sbjct 23 ESHKKQAKLILKRLSPTSEKRASIESGDYTFHYLIDNGICYLCICEQSYPRKLAFSYLEE 82
Query 71 VYLQFTQS 78
+ +F S
Sbjct 83 LAGEFWNS 90
> SPBC3F6.01c
Length=473
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Query 13 GLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPN 47
GLK V+++ R + EVEH+G C VF PN
Sbjct 396 GLKAVIRSHEVR---DQGYEVEHDGYCITVFSAPN 427
> At2g32830
Length=542
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 25/56 (44%), Gaps = 7/56 (12%)
Query 40 AFVFRYPNSLAALAVGDKEYPRRVA-------FACLNEVYLQFTQSVPASKWKSLE 88
AF F Y + D YP + AC+N + + FT VP SK KSLE
Sbjct 458 AFGFLYAAQSSDSEKTDAGYPPGIGVRNSLLMLACVNFLGIVFTLLVPESKGKSLE 513
> Hs22041471
Length=605
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 4/45 (8%)
Query 4 FYXRNTVKEGLKFVVKTAIPRLPV----NSQQEVEHEGLCAFVFR 44
FY + TV +G + IPR+ V N+ +E +H L ++FR
Sbjct 384 FYHQGTVVKGSFLISVVRIPRIIVMYMQNALKEQQHGALSRYLFR 428
Lambda K H
0.322 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40