bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4976_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
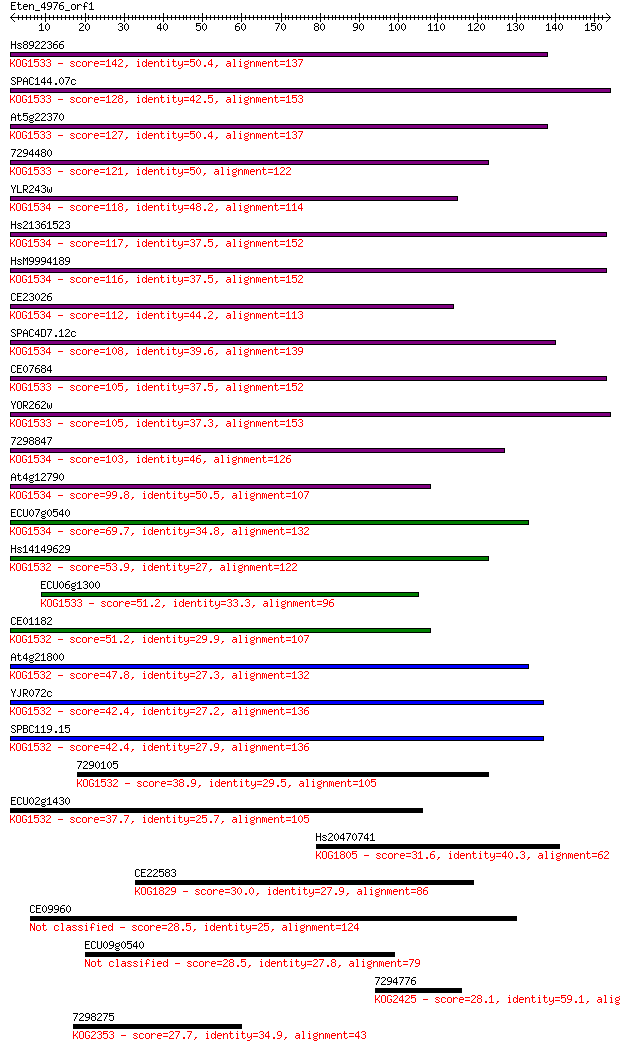
Hs8922366 142 3e-34
SPAC144.07c 128 4e-30
At5g22370 127 7e-30
7294480 121 6e-28
YLR243w 118 4e-27
Hs21361523 117 1e-26
HsM9994189 116 2e-26
CE23026 112 3e-25
SPAC4D7.12c 108 4e-24
CE07684 105 3e-23
YOR262w 105 5e-23
7298847 103 2e-22
At4g12790 99.8 2e-21
ECU07g0540 69.7 2e-12
Hs14149629 53.9 1e-07
ECU06g1300 51.2 7e-07
CE01182 51.2 8e-07
At4g21800 47.8 9e-06
YJR072c 42.4 3e-04
SPBC119.15 42.4 4e-04
7290105 38.9 0.004
ECU02g1430 37.7 0.009
Hs20470741 31.6 0.56
CE22583 30.0 1.7
CE09960 28.5 4.7
ECU09g0540 28.5 5.2
7294776 28.1 6.1
7298275 27.7 9.9
> Hs8922366
Length=310
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/137 (50%), Positives = 88/137 (64%), Gaps = 1/137 (0%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NG ++YCMEYL N+DWL KL+ + HY L DCPGQVEL THH ALRSI + D R
Sbjct 78 NGGLLYCMEYLEANLDWLRAKLDPLRGHYFLFDCPGQVELCTHHGALRSIFSQMAQWDLR 137
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFY 120
+ AV +VDS CTD K IS L +L L +ELPH+N+LSK+DL+ H L F L++Y
Sbjct 138 LTAVHLVDSHYCTDPAKFISVLCTSLATMLHVELPHINLLSKMDLIE-HYGKLAFNLDYY 196
Query 121 ADAYELQPLLQAVQEDP 137
+ +L LL + DP
Sbjct 197 TEVLDLSYLLDHLASDP 213
> SPAC144.07c
Length=315
Score = 128 bits (322), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 104/154 (67%), Gaps = 4/154 (2%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLES-LDF 59
NGA+IY ME + +++WL+++L ++ YV+ DCPGQVELFT+H++L+ II+ LE LD+
Sbjct 71 NGALIYAMEAIEYHVEWLLKELKKHRDSYVIFDCPGQVELFTNHNSLQKIIKTLEKELDY 130
Query 60 RMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEF 119
R +VQ+VD+ CT+ ++SALL+ L LQL++PHVN+LSK DLL + L +L+F
Sbjct 131 RPVSVQLVDAYCCTNPSAYVSALLVCLKGMLQLDMPHVNILSKADLLCTYGT-LPMKLDF 189
Query 120 YADAYELQPLLQAVQEDPHPLGRKLMDFSRAVCE 153
+ + +L L + D ++ D ++A+CE
Sbjct 190 FTEVQDLSYLAPLLDRDKRL--QRYSDLNKAICE 221
> At5g22370
Length=291
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 98/139 (70%), Gaps = 3/139 (2%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAF-KNHYVLIDCPGQVELFTHHDALRSIIRHL-ESLD 58
NG ++YCMEYL +NIDWL KL K+HY+L D PGQVELF HD+ ++++ L +SL+
Sbjct 71 NGGLVYCMEYLEKNIDWLESKLKPLLKDHYILFDFPGQVELFFIHDSTKNVLTKLIKSLN 130
Query 59 FRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLE 118
R+ AVQ++DS LC D ++S+LL++L+ L +ELPHVNVLSK+DL+ + K L F L+
Sbjct 131 LRLTAVQLIDSHLCCDPGNYVSSLLLSLSTMLHMELPHVNVLSKIDLIGSYGK-LAFNLD 189
Query 119 FYADAYELQPLLQAVQEDP 137
FY D +L L + +DP
Sbjct 190 FYTDVQDLSYLEHHLSQDP 208
> 7294480
Length=306
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 61/126 (48%), Positives = 85/126 (67%), Gaps = 5/126 (3%)
Query 1 NGAMIYCMEYLLENI-DWLV---EKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLES 56
NGA+++C EYL +++ DWL+ KL+A N Y L DCPGQVEL+THH+A+ I LE
Sbjct 83 NGALMHCAEYLADHLEDWLLPALRKLSATYN-YFLFDCPGQVELYTHHNAMARIFERLER 141
Query 57 LDFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFR 116
+ + V ++DS C++ K I+ LLMALN L++ LPHVNVLSK DLL+ H L F
Sbjct 142 ERYSLVTVNLIDSHYCSEPAKFIATLLMALNTMLRMSLPHVNVLSKADLLKKHETKLHFN 201
Query 117 LEFYAD 122
+++Y D
Sbjct 202 VDYYTD 207
> YLR243w
Length=272
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 55/117 (47%), Positives = 85/117 (72%), Gaps = 3/117 (2%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHL-ESLDF 59
NGA+IYC EYLL+N+DWL E++ F + Y++ DCPGQ+EL+TH L +I+RHL + L+F
Sbjct 72 NGALIYCFEYLLKNLDWLDEEIGDFNDEYLIFDCPGQIELYTHIPVLPNIVRHLTQQLNF 131
Query 60 RMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLR--LHRKDLK 114
+ A ++++ D+ K S L A++A + LELPH+NVLSK+DL++ +++K LK
Sbjct 132 NLCATYLLEAPFVIDSSKFFSGALSAMSAMILLELPHINVLSKLDLIKGDINKKKLK 188
> Hs21361523
Length=284
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 57/152 (37%), Positives = 92/152 (60%), Gaps = 6/152 (3%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NG +++CMEY N DWL L ++ Y+L DCPGQ+EL+TH ++ +++ LE +FR
Sbjct 74 NGGLVFCMEYFANNFDWLENCLGHVEDDYILFDCPGQIELYTHLPVMKQLVQQLEQWEFR 133
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFY 120
+ V +VDS ++FK IS +L AL+A + LE+P VN+++K+DLL K K +E +
Sbjct 134 VCGVFLVDSQFMVESFKFISGILAALSAMISLEIPQVNIMTKMDLL---SKKAKKEIEKF 190
Query 121 ADAYELQPLLQAVQEDPHPLGRKLMDFSRAVC 152
D ++ LL+ D +K ++A+C
Sbjct 191 LDP-DMYSLLEDSTSDLR--SKKFKKLTKAIC 219
> HsM9994189
Length=284
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 57/152 (37%), Positives = 92/152 (60%), Gaps = 6/152 (3%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NG +++CMEY N DWL L ++ Y+L DCPGQ+EL+TH ++ +++ LE +FR
Sbjct 74 NGGLVFCMEYFANNFDWLENCLGHVEDDYILFDCPGQIELYTHLPVMKQLVQQLEQWEFR 133
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFY 120
+ V +VDS ++FK IS +L AL+A + LE+P VN+++K+DLL K K +E +
Sbjct 134 VCGVFLVDSQFMVESFKFISGILAALSAMISLEIPQVNIMTKMDLL---SKKAKKEIEKF 190
Query 121 ADAYELQPLLQAVQEDPHPLGRKLMDFSRAVC 152
D ++ LL+ D +K ++A+C
Sbjct 191 LDP-DMYSLLEDSTSDLR--SKKFKKLTKAIC 219
> CE23026
Length=272
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 50/113 (44%), Positives = 80/113 (70%), Gaps = 0/113 (0%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NGA+++CMEYL++N++WL ++L+ ++ Y +IDCPGQ+EL++H +R I+ L+S DF
Sbjct 73 NGALVFCMEYLVQNLEWLHDELDEGEDDYFVIDCPGQIELYSHLPVMRQIVDALKSWDFN 132
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDL 113
+ +V ++D+ DA K IS L AL+A + +E P +NVL+K+DLL K L
Sbjct 133 VCSVFLIDTNFVLDAEKFISGALTALSAMVAIETPAINVLTKMDLLSERNKQL 185
> SPAC4D7.12c
Length=276
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 91/142 (64%), Gaps = 6/142 (4%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLE-SLDF 59
NG +IYC E+L+EN+DWL E++ + Y++ D PGQ+EL+TH L ++IRHL+ +L+F
Sbjct 72 NGGLIYCFEFLMENLDWLNEEIGDYDEDYLIFDMPGQIELYTHVPILPALIRHLQVTLNF 131
Query 60 RMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEF 119
R AV +++S D K + +L A++A + +E+PH+N+LSK+DLL+ + K L+
Sbjct 132 RPCAVYLLESQFLVDRTKFFAGVLSAMSAMVMMEVPHINLLSKMDLLKDNNNITKAELKR 191
Query 120 YADAYELQPLL--QAVQEDPHP 139
+ + PLL + E +P
Sbjct 192 FLNT---DPLLLTGEINETTNP 210
> CE07684
Length=300
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 57/152 (37%), Positives = 84/152 (55%), Gaps = 5/152 (3%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NGA+ YC+E L N +WL++K+ A Y++IDCPGQ+EL+ L +IR LE R
Sbjct 102 NGALKYCIETLGANCNWLLQKIEANHKKYLIIDCPGQLELYKSEGELWKVIRFLEKSGVR 161
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFY 120
+ A+ + DS C+D K IS L L + +E+P VN LSK DL +D + LEF+
Sbjct 162 LCALHLADSLYCSDPSKFISVALSTLATMVTMEMPQVNCLSKADLF---SEDGTYDLEFF 218
Query 121 ADAYELQPLLQAVQEDPHPLGRKLMDFSRAVC 152
+ ++ LL + E P K + A+C
Sbjct 219 SHLPDVNRLLDLLNEVPGL--EKYRKLNEAIC 248
> YOR262w
Length=347
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 57/160 (35%), Positives = 97/160 (60%), Gaps = 10/160 (6%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAF---KNHYVLIDCPGQVELFTHHDALRSIIRHLES- 56
NG ++Y +E L +ID + ++ + + Y++ DCPGQVELFTHH +L +I + +E
Sbjct 71 NGGLMYAVESLDNSIDLFILQIKSLVEEEKAYLVFDCPGQVELFTHHSSLFNIFKKMEKE 130
Query 57 LDFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFR 116
LD R V ++D T ++IS LL+AL + L ++LPH+NV SK+D+L+ + +L FR
Sbjct 131 LDIRFCVVNLIDCFYMTSPSQYISILLLALRSMLMMDLPHINVFSKIDMLKSY-GELPFR 189
Query 117 LEFYADAYE---LQPLLQAVQEDPHPLGRKLMDFSRAVCE 153
L++Y + + L+P ++ +E LG+K + + E
Sbjct 190 LDYYTEVQDLDYLEPYIE--KEGSSVLGKKYSKLTETIKE 227
> 7298847
Length=235
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 58/136 (42%), Positives = 79/136 (58%), Gaps = 12/136 (8%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNH---------YVLIDCPGQVELFTHHDALRSII 51
NG +I+C+E+L+EN +WL E+L +N Y+L D PGQ+ELFTH R ++
Sbjct 73 NGGLIFCLEFLIENQEWLKEQLCGGENELMVGEPDDDYILFDMPGQIELFTHLKMGRQLV 132
Query 52 RHLESLDFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLH-R 110
LES +FR V +DS D K IS + AL+ +E PH+NVL+KVDLL R
Sbjct 133 ELLESWNFRTCVVFCLDSQFMVDGAKFISGTMAALSVMANMEQPHINVLTKVDLLSSDAR 192
Query 111 KDLKFRLEFYADAYEL 126
K L+ LE DA+ L
Sbjct 193 KQLEMYLE--PDAHSL 206
> At4g12790
Length=282
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/136 (39%), Positives = 79/136 (58%), Gaps = 29/136 (21%)
Query 1 NGAMIYCMEY-------LLENI---------------------DWLVEKLNAFKNH-YVL 31
NGA++YCMEY LEN+ DW+ E+L +++ Y++
Sbjct 71 NGALMYCMEYPLFWLHWQLENVTSFVKSGLEKLLTLYLEDSLHDWVDEELENYRDDDYLI 130
Query 32 IDCPGQVELFTHHDALRSIIRHLESLDFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQ 91
DCPGQ+ELFTH L++ + HL+ +F + V ++DS TD K IS + +L A +Q
Sbjct 131 FDCPGQIELFTHVPVLKNFVEHLKQKNFNVCVVYLLDSQFITDVTKFISGCMSSLAAMIQ 190
Query 92 LELPHVNVLSKVDLLR 107
LELPHVN+LSK+DLL+
Sbjct 191 LELPHVNILSKMDLLQ 206
> ECU07g0540
Length=252
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 75/132 (56%), Gaps = 8/132 (6%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NG ++ +E L ENI+ L L + +++ DCPGQ+ELF H D + II H+ F+
Sbjct 71 NGGLLLALEELYENIEEL--GLEDLEGSFLVFDCPGQIELFMHSDVMPKIIEHVGRY-FK 127
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFY 120
V +++S D K++S AL + +L++P +NV+SK+DL++ +DL+ FY
Sbjct 128 CGVVYVMESQYLVDINKYVSGCFCALISMARLDVPCINVISKMDLIK--NEDLEV---FY 182
Query 121 ADAYELQPLLQA 132
EL L+ A
Sbjct 183 TPTEELSMLIGA 194
> Hs14149629
Length=374
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 63/124 (50%), Gaps = 8/124 (6%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKN--HYVLIDCPGQVELFTHHDALRSIIRHLESLD 58
NG ++ + D +++ + +N YVLID PGQ+E+FT A +II +
Sbjct 88 NGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVLIDTPGQIEVFT-WSASGTIITEALASS 146
Query 59 FRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLE 118
F + ++D++ T+ +S +L A + + +LP + V++K D++ D F +E
Sbjct 147 FPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFIVVMNKTDII-----DHSFAVE 201
Query 119 FYAD 122
+ D
Sbjct 202 WMQD 205
> ECU06g1300
Length=266
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 47/96 (48%), Gaps = 8/96 (8%)
Query 9 EYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFRMAAVQIVD 68
E+ ENI+ +HYVL D PGQVE F D I+R+L S + + V + D
Sbjct 84 EFFRENIEN--------TDHYVLFDFPGQVEFFMSSDIPNGILRYLRSNGYSVVVVNLTD 135
Query 69 STLCTDAFKHISALLMALNAQLQLELPHVNVLSKVD 104
++ +S+ L++ LE VNV+SK D
Sbjct 136 LVFFSNDHSLLSSYLVSTLCLCLLESAQVNVISKCD 171
> CE01182
Length=355
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 56/109 (51%), Gaps = 3/109 (2%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHY--VLIDCPGQVELFTHHDALRSIIRHLESLD 58
NGA++ C+ + D ++E +N + + L+D PGQ+E FT A SII +
Sbjct 99 NGAIMTCLNLMCTRFDKVIELINKRSSDFSVCLLDTPGQIEAFTWS-ASGSIITDSLASS 157
Query 59 FRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLR 107
+ IVDS T+ +S +L A + + +LP + V +K D+++
Sbjct 158 HPTVVMYIVDSARATNPTTFMSNMLYACSILYRTKLPFIVVFNKADIVK 206
> At4g21800
Length=379
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 59/134 (44%), Gaps = 8/134 (5%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNH--YVLIDCPGQVELFTHHDALRSIIRHLESLD 58
NG ++ + D +V + + YVL+D PGQ+E+FT A +II +
Sbjct 110 NGGILTSLNLFATKFDEVVSVIEKRADQLDYVLVDTPGQIEIFTWS-ASGAIITEAFAST 168
Query 59 FRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLE 118
F +VD+ + +S +L A + + LP V +K D+ D KF LE
Sbjct 169 FPTVVTYVVDTPRSSSPITFMSNMLYACSILYKTRLPLVLAFNKTDV-----ADHKFALE 223
Query 119 FYADAYELQPLLQA 132
+ D Q +Q+
Sbjct 224 WMEDFEVFQAAIQS 237
> YJR072c
Length=385
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 63/140 (45%), Gaps = 15/140 (10%)
Query 1 NGAMIYCMEYLLENIDWLV----EKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLES 56
NGA++ + ID ++ +K + F+N +ID PGQ+E F A +II +
Sbjct 72 NGAIVTSLNLFSTKIDQVIRLVEQKKDKFQN--CIIDTPGQIECFVWS-ASGAIITESFA 128
Query 57 LDFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFR 116
F IVD+ + +S +L A + + +LP + V +K D+ + F
Sbjct 129 SSFPTVIAYIVDTPRNSSPTTFMSNMLYACSILYKTKLPMIVVFNKTDVCKAD-----FA 183
Query 117 LEFYADAYELQPLLQAVQED 136
E+ D Q A++ED
Sbjct 184 KEWMTDFESFQA---AIKED 200
> SPBC119.15
Length=367
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 66/139 (47%), Gaps = 13/139 (9%)
Query 1 NGAMIYCMEYLLENIDW---LVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESL 57
NG ++ + + D ++EK +H +LID PGQ+E+F A SII +
Sbjct 77 NGGIMTSLNLFVTKFDQVLKILEKRAPTVDH-ILIDTPGQIEIF-QWSASGSIICDTLAS 134
Query 58 DFRMAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRL 117
+ +VD+ T +S++L A + + +LP + V +K D+ +D +F
Sbjct 135 SWPTCIAYVVDTPRATSTSTWMSSMLYACSMLYKAKLPLIIVYNKCDV-----QDSEFAK 189
Query 118 EFYADAYELQPLLQAVQED 136
++ D E Q QAV +D
Sbjct 190 KWMTDFEEFQ---QAVTKD 205
> 7290105
Length=382
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 52/107 (48%), Gaps = 10/107 (9%)
Query 18 LVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFRMAAVQIVD--STLCTDA 75
LV + + + +ID PGQ+E+F + A SII + F V ++D + C
Sbjct 111 LVRRAGERGHKWCVIDTPGQIEVF-NWSASGSIITEGLATMFPTIVVYVMDVERSACPTT 169
Query 76 FKHISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFYAD 122
F +S +L A + + LP + L+K+DL KD F +++ D
Sbjct 170 F--MSNMLYACSILYKTRLPFLVALNKIDL-----KDCGFVMDWMTD 209
> ECU02g1430
Length=270
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 50/105 (47%), Gaps = 4/105 (3%)
Query 1 NGAMIYCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFR 60
NG + C+ L NI ++++ YV++D PGQ+E FT +I L+++
Sbjct 95 NGGITTCLNLFLLNIGTYIDRI---VEEYVIVDTPGQIEAFTWSSPGYVLIETLKTIG-D 150
Query 61 MAAVQIVDSTLCTDAFKHISALLMALNAQLQLELPHVNVLSKVDL 105
+ V VDS +S ++ A + + E+ + + +K DL
Sbjct 151 VILVYTVDSLSSHKHAVFMSNMMYAASLMCRYEVETLCLFNKKDL 195
> Hs20470741
Length=1060
Score = 31.6 bits (70), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 79 ISALLMALNAQLQLELPHVNVLSKVDLLRLHRKDLKFRLEFYADAYELQPLLQAVQEDPH 138
I +L L + +LE V + V LR H KD+K LEFYAD Y P L V E +
Sbjct 833 IMSLSNKLTYEGKLECGSDKVANAVINLR-HFKDVKLELEFYAD-YSDNPWLMGVFEPNN 890
Query 139 PL 140
P+
Sbjct 891 PV 892
> CE22583
Length=619
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 45/87 (51%), Gaps = 6/87 (6%)
Query 33 DCPGQVELFTHHDALRSIIRHLESLDFRMAAVQI-VDSTLCTDAFKHISALLMALNAQLQ 91
D PG +L + ALR+++ + L ++I +D++ F ++S + +++ L
Sbjct 422 DLPG--DLVKKNKALRAVVELRQKLKHMEGFIKICIDASNQVFEFGNLSTMFASIDRYL- 478
Query 92 LELPHVNVLSKVDLLRLHRKDLKFRLE 118
L H ++ S DL R++ KDL LE
Sbjct 479 --LEHDDLFSLNDLQRIYNKDLLSLLE 503
> CE09960
Length=843
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 31/141 (21%), Positives = 61/141 (43%), Gaps = 17/141 (12%)
Query 6 YCMEYLLENIDWLVEKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESL--DFRMAA 63
+ + Y +N+ V F N VL P ++ +F +HD + + D R +
Sbjct 21 FSLTYAAKNLSQFVAGEKQFSNEKVLFGIPWRLVVFENHDEIVGLSLSCGQCVQDSRKWS 80
Query 64 VQI-VDSTLCTDA---FKHISALLMALNAQLQLELPHV-----------NVLSKVDLLRL 108
+ I V+ L + + F + LL+ +++L+L +V +VD+ L
Sbjct 81 LSIEVELKLMSSSEKGFSKVRRLLVKTLTKIELQLLDTEDLIENFVSAESVRIQVDVKIL 140
Query 109 HRKDLKFRLEFYADAYELQPL 129
+K+LK+ + + Y +Q L
Sbjct 141 EQKELKYETKIFELTYPIQNL 161
> ECU09g0540
Length=567
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 38/85 (44%), Gaps = 11/85 (12%)
Query 20 EKLNAFKNHYVLIDCPGQVELFTHHDALRSIIRHLESLDFRMAAVQ------IVDSTLCT 73
E LN+F +Y+ + PG+ E +AL+ ++ +++L ++ IVD
Sbjct 40 ESLNSFLGYYLNREIPGREECVAAKEALKILMDGIKALIPSYYTIEAESFFKIVD----- 94
Query 74 DAFKHISALLMALNAQLQLELPHVN 98
D KH L L+ LP+ N
Sbjct 95 DLIKHSKESLQVFKNALREALPYAN 119
> 7294776
Length=550
Score = 28.1 bits (61), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 17/23 (73%), Gaps = 1/23 (4%)
Query 94 LPHVNVLSKVDLLRLHRKD-LKF 115
LPH+ +S +D +LHRKD LKF
Sbjct 185 LPHMETMSNLDAGKLHRKDKLKF 207
> 7298275
Length=2172
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 3/46 (6%)
Query 17 WLVEKLNAFKNHYVLIDCPGQVELFTHHDA---LRSIIRHLESLDF 59
W +E K+ +L+D G + F HH A +RSI+ + DF
Sbjct 215 WYIETATCSKDIVILLDHSGSMTGFRHHVAKFTIRSILDTFSNNDF 260
Lambda K H
0.327 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40