bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4929_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
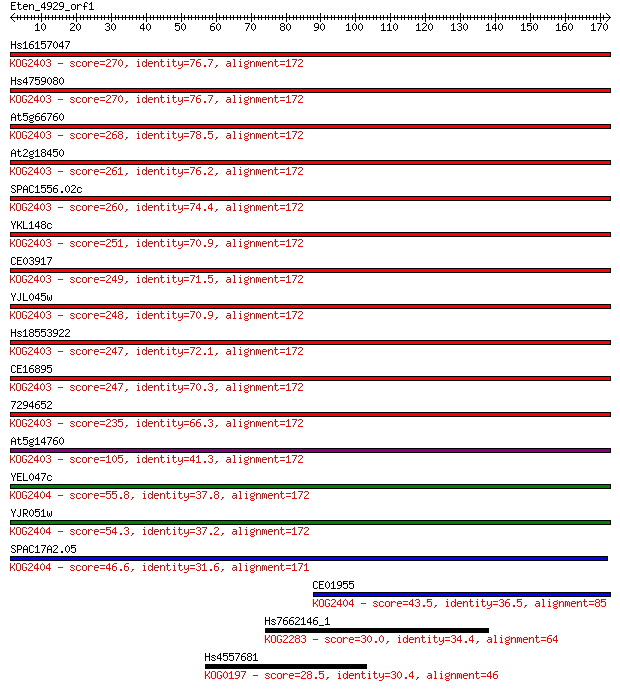
Hs16157047 270 8e-73
Hs4759080 270 9e-73
At5g66760 268 3e-72
At2g18450 261 5e-70
SPAC1556.02c 260 1e-69
YKL148c 251 4e-67
CE03917 249 2e-66
YJL045w 248 4e-66
Hs18553922 247 7e-66
CE16895 247 1e-65
7294652 235 4e-62
At5g14760 105 4e-23
YEL047c 55.8 4e-08
YJR051w 54.3 1e-07
SPAC17A2.05 46.6 2e-05
CE01955 43.5 2e-04
Hs7662146_1 30.0 2.5
Hs4557681 28.5 6.3
> Hs16157047
Length=664
Score = 270 bits (691), Expect = 8e-73, Method: Compositional matrix adjust.
Identities = 132/172 (76%), Positives = 147/172 (85%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NS+GE FM RYAP AKDLASRDVVSRSMT+EIR
Sbjct 293 VQFHPTGIYGAGCLITEGCRGEGGILINSQGERFMERYAPVAKDLASRDVVSRSMTLEIR 352
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +L L HL PE L +RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 353 EGRGCGPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 412
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +V+ V G+ + ++ GLYA GEAACASVHGANRLGANSLLD
Sbjct 413 IPTNYKGQVLRHVNGQDQ-----IVPGLYACGEAACASVHGANRLGANSLLD 459
> Hs4759080
Length=664
Score = 270 bits (691), Expect = 9e-73, Method: Compositional matrix adjust.
Identities = 132/172 (76%), Positives = 147/172 (85%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NS+GE FM RYAP AKDLASRDVVSRSMT+EIR
Sbjct 293 VQFHPTGIYGAGCLITEGCRGEGGILINSQGERFMERYAPVAKDLASRDVVSRSMTLEIR 352
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +L L HL PE L +RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 353 EGRGCGPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 412
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +V+ V G+ + ++ GLYA GEAACASVHGANRLGANSLLD
Sbjct 413 IPTNYKGQVLRHVNGQDQ-----IVPGLYACGEAACASVHGANRLGANSLLD 459
> At5g66760
Length=634
Score = 268 bits (686), Expect = 3e-72, Method: Compositional matrix adjust.
Identities = 135/172 (78%), Positives = 145/172 (84%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGGILRNSEGE FM RYAPTAKDLASRDVVSRSMT+EIR
Sbjct 282 VQFHPTGIYGAGCLITEGSRGEGGILRNSEGERFMERYAPTAKDLASRDVVSRSMTMEIR 341
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL PE L RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 342 EGRGVGPHKDHIYLHLNHLPPEVLKERLPGISETAAIFAGVDVTKEPIPVLPTVHYNMGG 401
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN+ EV+ +KG + V+ GL AAGEAACASVHGANRLGANSLLD
Sbjct 402 IPTNYHGEVV-TIKGDD---PDAVIPGLMAAGEAACASVHGANRLGANSLLD 449
> At2g18450
Length=632
Score = 261 bits (667), Expect = 5e-70, Method: Compositional matrix adjust.
Identities = 131/172 (76%), Positives = 145/172 (84%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGGILRNSEGE FM RYAPTA+DLASRDVVSRSMT+EIR
Sbjct 280 VQFHPTGIYGAGCLITEGARGEGGILRNSEGEKFMDRYAPTARDLASRDVVSRSMTMEIR 339
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
+GRG GP KD+ +L L HL PE L RLPGI+ETA IFAGVDVT++PIPVLPTVHYNMGG
Sbjct 340 QGRGAGPMKDYLYLYLNHLPPEVLKERLPGISETAAIFAGVDVTREPIPVLPTVHYNMGG 399
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN+ EVI ++G + V+ GL AAGEAACASVHGANRLGANSLLD
Sbjct 400 IPTNYHGEVI-TLRGDD---PDAVVPGLMAAGEAACASVHGANRLGANSLLD 447
> SPAC1556.02c
Length=641
Score = 260 bits (664), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 128/172 (74%), Positives = 143/172 (83%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGG L NS+GE FM RYAPTAKDLASRDVVSR+MT+EIR
Sbjct 286 VQFHPTGIYGAGCLITEGCRGEGGYLLNSKGERFMERYAPTAKDLASRDVVSRAMTVEIR 345
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDHC+L L+HL E L RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 346 EGRGVGPEKDHCYLQLSHLPAEILKERLPGISETAAIFAGVDVTKEPIPVLPTVHYNMGG 405
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT + EV+ + G+ + ++ GLYAAGEAAC SVHG NRLGANSLLD
Sbjct 406 IPTRFTGEVL-TIDENGK---DKIVPGLYAAGEAACVSVHGGNRLGANSLLD 453
> YKL148c
Length=640
Score = 251 bits (642), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 122/172 (70%), Positives = 139/172 (80%), Gaps = 3/172 (1%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHP+GI+ +GCLITEG RGEGG L NSEGE FM RYAPTAKDLA RDVVSR++T+EIR
Sbjct 284 VQFHPSGIYGSGCLITEGARGEGGFLVNSEGERFMERYAPTAKDLACRDVVSRAITMEIR 343
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG G +KDH +L L+HL PE L RLPGI+ETA IFAGVDVTK+PIP++PTVHYNMGG
Sbjct 344 EGRGVGKKKDHMYLQLSHLPPEVLKERLPGISETAAIFAGVDVTKEPIPIIPTVHYNMGG 403
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT W E + + G E+ V+ GL A GEAAC SVHGANRLGANSLLD
Sbjct 404 IPTKWNGEALTIDEETG---EDKVIPGLMACGEAACVSVHGANRLGANSLLD 452
> CE03917
Length=646
Score = 249 bits (635), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 123/172 (71%), Positives = 137/172 (79%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGG L NS GE FM RYAP AKDLASRDVVSRSMT+EI
Sbjct 274 VQFHPTGIYGAGCLITEGSRGEGGYLVNSAGERFMERYAPNAKDLASRDVVSRSMTVEIM 333
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL E L RLPGI+ETA IFAGVDVTK+PIPV+PTVHYNMGG
Sbjct 334 EGRGVGPDKDHIYLQLHHLPAEQLQQRLPGISETAMIFAGVDVTKEPIPVIPTVHYNMGG 393
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
+PTN++ +V++ KG + V+ GLYAAGE SVHGANRLGANSLLD
Sbjct 394 VPTNYKGQVLNYTPKKG----DEVVPGLYAAGECGAHSVHGANRLGANSLLD 441
> YJL045w
Length=634
Score = 248 bits (633), Expect = 4e-66, Method: Compositional matrix adjust.
Identities = 122/172 (70%), Positives = 137/172 (79%), Gaps = 3/172 (1%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHP+GI+ +GCLITEG RGEGG L NSEGE FM RYAPTAKDLASRDVVSR++T+EIR
Sbjct 278 VQFHPSGIYGSGCLITEGARGEGGFLLNSEGERFMERYAPTAKDLASRDVVSRAITMEIR 337
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
GRG G KDH L L+HL PE L RLPGI+ETA +FAGVDVT++PIPVLPTVHYNMGG
Sbjct 338 AGRGVGKNKDHILLQLSHLPPEVLKERLPGISETAAVFAGVDVTQEPIPVLPTVHYNMGG 397
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT W E + + G E+ V+ GL A GEAAC SVHGANRLGANSLLD
Sbjct 398 IPTKWTGEALTIDEETG---EDKVIPGLMACGEAACVSVHGANRLGANSLLD 446
> Hs18553922
Length=365
Score = 247 bits (631), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 124/172 (72%), Positives = 138/172 (80%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTG + AGCLITEGCRGEGGIL NS+GE FM RYAP AKDLASRDVVSR MT+EIR
Sbjct 180 VQFHPTGTYGAGCLITEGCRGEGGILINSQGERFMERYAPIAKDLASRDVVSRWMTLEIR 239
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +L L HL PE L LPGI+ETA IFAGVDVTK+PIPVLPTVHYNM G
Sbjct 240 EGRGCGPEKDHVYLQLHHLPPEQLAMPLPGISETAMIFAGVDVTKEPIPVLPTVHYNMDG 299
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT++ +V+ G+ + ++ GLYA GEAACAS HG NRLGANSLLD
Sbjct 300 IPTSYEGQVLRHGNGQDQ-----IVPGLYACGEAACASAHGVNRLGANSLLD 346
> CE16895
Length=640
Score = 247 bits (630), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 121/172 (70%), Positives = 137/172 (79%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ GCLITEG RGEGG L NS+GE FM RYAP AKDLASRDVVSR+MT+EI
Sbjct 267 VQFHPTGIYGVGCLITEGSRGEGGYLVNSQGERFMERYAPNAKDLASRDVVSRAMTMEIN 326
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL E L RLPGI+ETA+IFAGVDVTK+PIPV+PTVHYNMGG
Sbjct 327 EGRGVGPNKDHIYLQLHHLPAEQLQQRLPGISETAQIFAGVDVTKEPIPVIPTVHYNMGG 386
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
+PTN++ +V+ G + V+ GLYAAGE A SVHGANRLGANSLLD
Sbjct 387 VPTNYKGQVLDFTPEGG----DKVIPGLYAAGECAAHSVHGANRLGANSLLD 434
> 7294652
Length=525
Score = 235 bits (599), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 114/172 (66%), Positives = 136/172 (79%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGG N +GE FM RYAP AKDLASRDVV+R+MT+E+
Sbjct 154 VQFHPTGIYGAGCLITEGVRGEGGFFLNCKGERFMERYAPKAKDLASRDVVARAMTMEVL 213
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
G GCGP KDH HL L H+D + + RLPGI TA+IFA VDVTK+P+PVLPTVHYNMGG
Sbjct 214 AGNGCGPLKDHVHLQLHHIDAKIIKQRLPGIMVTARIFAKVDVTKEPVPVLPTVHYNMGG 273
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT+++ V+ + G+ E V++GLY+ GE +CASVHGANRLGANSLLD
Sbjct 274 IPTDYKGRVV-TIDENGK---EQVVKGLYSCGETSCASVHGANRLGANSLLD 321
> At5g14760
Length=642
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 96/184 (52%), Gaps = 33/184 (17%)
Query 1 VQFHPTGIFPAGC------------LITEGCRGEGGILRNSEGEAFMARYAPTAKDLASR 48
VQFHPT + G LITE RG+GGIL N E FM Y A +LA R
Sbjct 301 VQFHPTALADEGLPIKLQTARENAFLITEAVRGDGGILYNLGMERFMPVYDERA-ELAPR 359
Query 49 DVVSRSMTIEIREGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPI 108
DVV+RS+ ++++ + + LD++H E + + P I + G+D+T+QPI
Sbjct 360 DVVARSIDDQLKK-----RNEKYVLLDISHKPREKILAHFPNIASEC-LKHGLDITRQPI 413
Query 109 PVLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGAN 168
PV+P HY GG+ G E VL GL+ AGE AC +HGANRL +N
Sbjct 414 PVVPAAHYMCGGVRA-------------GLQGETNVL-GLFVAGEVACTGLHGANRLASN 459
Query 169 SLLD 172
SLL+
Sbjct 460 SLLE 463
> YEL047c
Length=470
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 65/223 (29%), Positives = 91/223 (40%), Gaps = 66/223 (29%)
Query 1 VQFHPTGIFPAG-------CLITEGCRGEGGILRNS-EGEAFMARYAPTAKDLASRDVVS 52
+Q HPTG L E RG GGIL N G F+ +L +RDVV+
Sbjct 246 IQVHPTGFIDPNDRSSSWKFLAAESLRGLGGILLNPITGRRFV-------NELTTRDVVT 298
Query 53 RSM--TIEIREGRGCGPRKDHCHLDL-THLDPETLHSRLPGIT----------------- 92
++ + R + + DL +LD + +T
Sbjct 299 AAIQKVCPQEDNRALLVMGEKMYTDLKNNLDFYMFKKLVQKLTLSQVVSEYNLPITVAQL 358
Query 93 ----ETAKIFA---------------GVDVTKQPI----PVLPTVHYNMGGIPTNWRSEV 129
+T F G DVT + + V P VH+ MGG N +++V
Sbjct 359 CEELQTYSSFTTKADPLGRTVILNEFGSDVTPETVVFIGEVTPVVHFTMGGARINVKAQV 418
Query 130 IHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
I GK +E +L+GLYAAGE + VHGANRLG +SLL+
Sbjct 419 I----GKN---DERLLKGLYAAGEVS-GGVHGANRLGGSSLLE 453
> YJR051w
Length=501
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 64/229 (27%), Positives = 93/229 (40%), Gaps = 78/229 (34%)
Query 1 VQFHPTGIFPAG-------CLITEGCRGEGGILRN-SEGEAFMARYAPTAKDLASRDVVS 52
VQ HPTG L E RG GGIL + + G F +L++RD V
Sbjct 278 VQVHPTGFIDPNDRENNWKFLAAEALRGLGGILLHPTTGRRF-------TNELSTRDTV- 329
Query 53 RSMTIEIR------EGRGCGPRKDHCHLDLTH----------------------LDPETL 84
T+EI+ + R D + + T+ D +T
Sbjct 330 ---TMEIQSKCPKNDNRALLVMSDKVYENYTNNINFYMSKNLIKKVSINDLIRQYDLQTT 386
Query 85 HSRLPGITETAKIFAGV---DVTKQPI------------------PVLPTVHYNMGGIPT 123
S L +TE K ++ V D +P+ V P VH+ MGG+
Sbjct 387 ASEL--VTE-LKSYSDVNTKDTFDRPLIINAFDKDISTESTVYVGEVTPVVHFTMGGVKI 443
Query 124 NWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
N +S+VI + +E + G++AAGE + VHGANRLG +SLL+
Sbjct 444 NEKSQVIK------KNSESVLSNGIFAAGEVS-GGVHGANRLGGSSLLE 485
> SPAC17A2.05
Length=513
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 54/207 (26%), Positives = 75/207 (36%), Gaps = 45/207 (21%)
Query 1 VQFHPTGIFP-------AGCLITEGCRGEGGILRNSEGEAF--------------MARYA 39
VQ HPTG L E RG G +L S+G F M +
Sbjct 285 VQIHPTGFVDPKDPTALTKFLAAEALRGSGAVLLTSQGRRFCDELGYRDYVTGEMMKLKS 344
Query 40 PTAKDLASRDVVSRSMTIEIREGRGCGPRKDHCHL-DLTHLDPETLHSRLPGITETAK-- 96
P L S + I+ +G + L + + L S AK
Sbjct 345 PVYLVLNSAAAEEVANFIKFYSFKGLMKKMKAEELCSTLNCTKDELASTFSEYNRAAKGE 404
Query 97 ---IFAGVDVTKQPIP---------VLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECV 144
F K P+ V+P +HY MGG+ + +S V+ V
Sbjct 405 IPDEFGRKYFGKTPLELTDTFTVGEVVPVLHYTMGGVQVDTQSRVLST--------NGNV 456
Query 145 LEGLYAAGEAACASVHGANRLGANSLL 171
++GL+AAGE +HG NRLG +SLL
Sbjct 457 IDGLFAAGE-IVGGIHGENRLGGSSLL 482
> CE01955
Length=474
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 44/85 (51%), Gaps = 17/85 (20%)
Query 88 LPGITETAKIFAGVDVTKQPIPVLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECVLEG 147
+ I+ T I+A + V+P +HY MGG+ + + VI GK + G
Sbjct 384 VSAISPTEPIYAAI--------VVPAIHYTMGGLKIDEATRVIDE-HGK-------PIVG 427
Query 148 LYAAGEAACASVHGANRLGANSLLD 172
L+AAGE VHG+NRL NSLL+
Sbjct 428 LFAAGEV-TGGVHGSNRLAGNSLLE 451
> Hs7662146_1
Length=648
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 10/73 (13%)
Query 74 LDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGGIPT---NWRSE-- 128
LD HLD T+++ P TAK + V PI P++H N+ + NW +
Sbjct 98 LDSRHLDHYTVYNLSPKSYRTAKFHSRVSECSWPIRQAPSLH-NLFAVCRNMYNWLLQNP 156
Query 129 ----VIHCVKGKG 137
V+HC+ G+
Sbjct 157 KNVCVVHCLDGRA 169
> Hs4557681
Length=1124
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 57 IEIREGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVD 102
I I++ GP +H + +T D + L + PG+ E A VD
Sbjct 301 ISIKQAPRVGPAGEHRLVTVTRTDNQILEAEFPGLPEALSFVALVD 346
Lambda K H
0.320 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40